Structural and Chemical Biology of Membrane Proteins

- Anirban Banerjee, PhD, Head, Unit on Structural and Chemical Biology of Membrane Proteins
- Eric Christenson, PhD, Postdoctoral Fellow
- Chul-jin Lee, PhD, Visiting Fellow
- Jin-Sik Kim, PhD, Visiting Fellow
- Robbins Puthenveetil, PhD, Visiting fellow
- Mitra Rana, PhD, Visiting Fellow
- Grace Wang, BS, Postbaccalaureate Fellow
Molecular mechanism of post-translational protein lipidation by DHHC palmitoyltransferases
Covalent modification of proteins by attachment of fatty acids to internal cysteines through acyl linkages constitutes one of the most pervasive and physiologically important post-translational modifications, which are referred to collectively as protein palmitoylation, or more generally, protein S-acylation. The repertoire of palmitoylated proteins has expanded rapidly in recent years, with hundreds of new members being added to the cellular "palmitoylome." The physico-chemical effect of palmitoylation is to alter the local hydrophobicity of the substrate protein. The thioester bond makes S-acylation unique in that it is a labile moiety and can be cleaved, in the cellular context, by thioesterase enzymes (Figure 1). Thus S-acylation is one of the few dynamic post-translational modifications. The physiological effects of S-acylation are diverse and have critical cellular importance; Ras, a small GTPase that is critical for cellular growth and differentiation and is mutated in about one-third of all human cancers, is palmitoylated at the Golgi and subsequently targeted to the plasma membrane by vesicular transport. Palmitoylated Ras localizes to cholesterol-rich domains on the plasma membrane. However, it is subsequently depalmitoylated by the thioesterase APT1, dissociates from the plasma membrane, and redistributes on endomembranes, including the Golgi. Such dynamic recycling of Ras is critical for its function.
Protein S-acylation is catalyzed by a large group of enzymes known as DHHC–palmitoyl acyltransferase (DHHC–PAT), so named because they contain a signature D-H-H-C motif in a cysteine-rich domain in an intracellular loop (Figure 1). These are low-abundance polytopic integral membrane proteins localized at a variety of cellular compartments. Yeast has seven DHHC–PATs and mammals up to 24 putative DHHC–PATs encoded in their genome. Beyond the shared DHHC domain, DHHC–PATs vary considerably; some possess ankyrin repeats (structural protein motifs that mediate protein-protein interactions), a few have six transmembrane helices instead of the usual four, and at least one forms a functional heterodimer with a cytoplasmic auxiliary subunit (Figure 1). To date, there are no reported consensus sequences for palmitoylation. A specific DHHC–PAT can palmitoylate many substrates, and, conversely, a given substrate can be palmitoylated by many DHHC–PATs. Such redundancy has been one of the most intriguing aspects of DHHC–PATs and makes it difficult to assign substrates by overexpression/knockout strategies, given that in the absence of one enzyme, others can take over. However, this does not necessarily reflect the true enzyme-substrate relationship. The situation has been even more confounded by the lack of specific inhibitors of DHHC–PATs. Even though 2-bromopalmitate is widely used as a global inhibitor of DHHC–PATs, it has been shown that it broadly targets other proteins involved in lipid metabolism.
Click image to view.
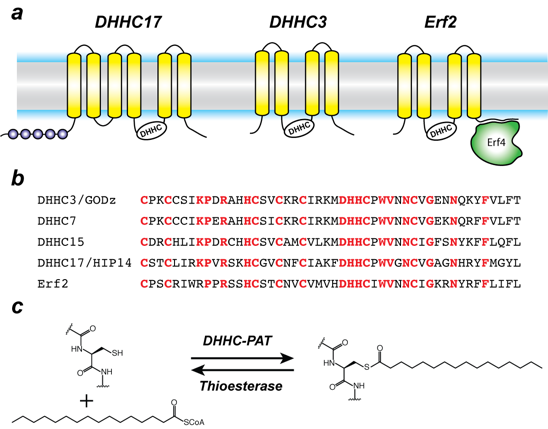
Figure 1. Organization and properties of DHHC palmitoyltransferases
a. The organization of three different DHHC-PATs are shown schematically. The spheres indicate protein-protein interaction domains. Erf2 associates with a cytoplasmic subunit, Erf4, to form the active enzyme.
b. The DHHC-CRD region of a few representative DHHC-PAT’s are aligned. The conserved amino acids are shown in red.
c. Reaction catalyzed by DHHC-PATs; the reverse reaction is catalyzed by acylprotein thioesterases (APT).
Besides its broad importance in cell biology, palmitoylation has been linked to several diseases, most notably neuro-psychiatric disorders such as Huntington’s disease and various forms of cancer. Very recently, it was shown that DHHC20 palmitoylates epidermal growth factor (EGFR) and is thus a potential therapeutic target for a broad range of cancers. However, despite their importance across a broad spectrum of biological pathways and their biomedical importance, very little is known about the molecular mechanism of DHHC palmitoyltransferases. There is nothing known about their structural organization or how they interact with substrates and the fatty acyl coenzyme A (CoA) that serves as the acyl donor.
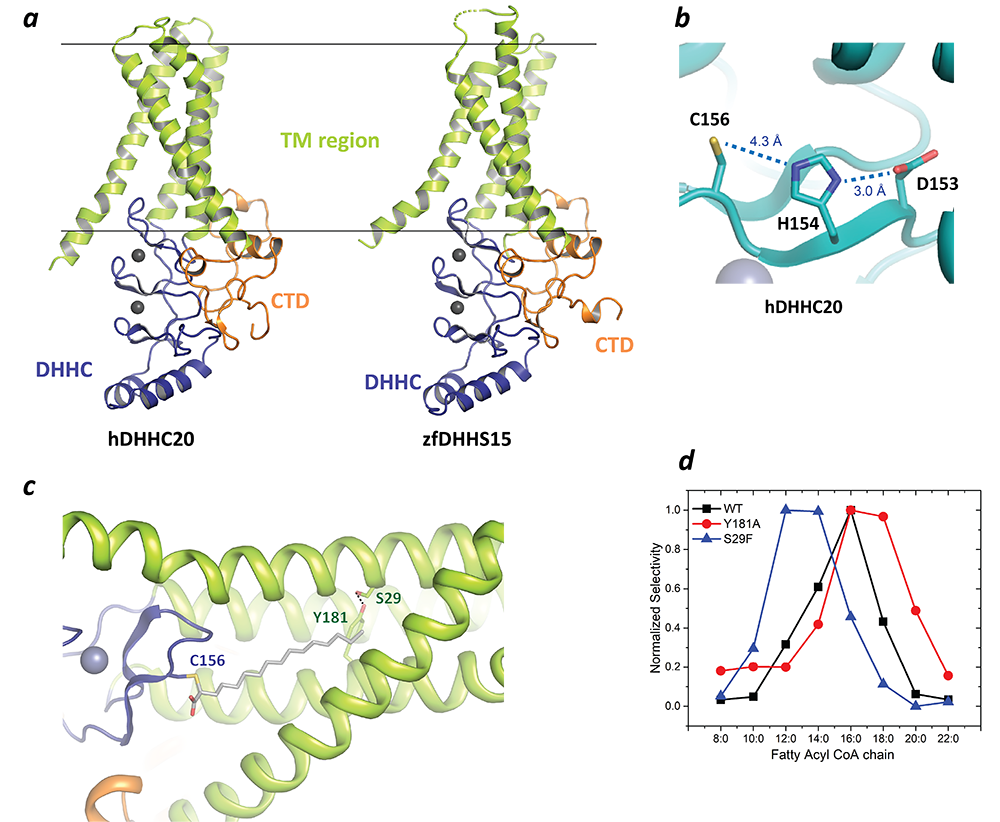
In a major breakthrough in this field, we solved the high-resolution crystal structures of two members of the DHHC family, human DHHC20 and zebrafish DHHC15 (Figure 3a). They are the first structures of any member of this family to be characterized and reveal a tepee-like transmembrane domain organization, which splays apart towards the cytoplasmic side and harbors the active site at the membrane-aqueous interfacial region (Figure 3b), thus readily explaining why membrane-proximal cysteines are palmitoylated. The structures also revealed that the tightly bound Zn2+ ions do not actively take part in catalysis but serve as structural Zn2+ ions. The C-terminus of DHHC–PATs is the least conserved overall but has two important conserved sequence motifs, and the structures show how these form critical points of contact with the DHHC–CRD (cysteine-rich domain) domain. Unexpectedly, the C-terminus has an amphipathic helix followed by a hydrophobic loop, which insert hydrophobic residues into the membrane and straddle the transmembrane domain, thus forming a supporting structure.
We also solved the structure of human DHHC20 irreversibly modified by a covalent inhibitor, 2-bromopalmitate. The structure mimics the auto-acylated intermediate state in the enzymatic pathway and thus reveals how the acyl group of fatty acyl–CoA binds in a cavity formed in the bilayer by the transmembrane domain (Figure 3c). Residues lining the cavity contact the acyl chain, and mutation of these residues affects enzymatic activity. By mutating two residues at the tapering end of the cavity, we also showed that we can change the acyl chain length selectivity of the mutant enzymes (Figure 3d). Thus, the cavity functions as a molecular ruler in determining the acyl chain length selectivity of human DHHC20. This is important because, although palmitate is the most prevalent fatty acid used by DHHC palmitoyltransferases, they can use fatty acyl–CoAs of other chain lengths, and this property varies between different members of the DHHC family.
Molecular mechanism of iron transport into mitochondria
The importance of iron in biology cannot be overstated. In higher organisms, mitochondria are the ‘hotspot’ for the cell biology of iron, because this is where Fe-S clusters are biosynthesized and iron is inserted into heme. Mitochondrial iron homeostasis plays a critical role in cellular iron homeostasis and in the overall physiology of the cell. In vertebrates, the only known major transporters of iron into mitochondria are mitoferrin-1 and mitoferrin-2, two homologous members of a large group of mitochondrial transporters known as the Mitochondrial Carrier family (Figures 2a,b). Mitoferrin-1 (Mfrn1) is expressed mainly in erythroid cells, while mitoferrin-2 is expressed ubiquitously. Knockout of Mfrn1 is embryonically lethal, reflecting the importance of mitoferrins in vertebrate physiology.
Click image to view.
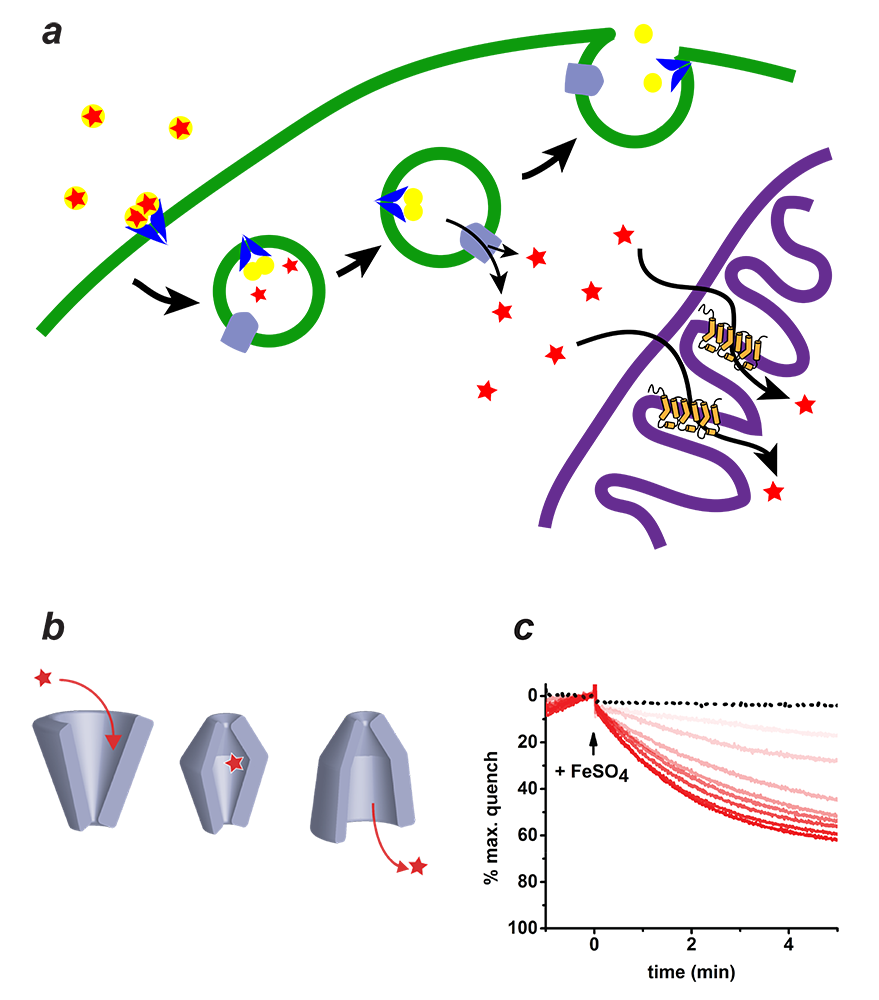
Figure 2. Mitoferrin and mitochondrial iron transport
a. Iron is imported through the plasma membrane by the Transferrin/Transferrin Receptor (blue) cycle and is transported out of endosomes by divalent metal ion transporter (DMT) (grey). The iron is delivered to Mitoferrin (yellow cylinders) by unknown means. Mitoferrin delivers iron to unknown partners in mitochondria, which become available for heme and Fe-S cluster biosynthesis.
b. Schematic depiction of the simplest transporter cycle and the different putative states of Mitoferrin involved.
c. Assay of iron transport activity by TMfrn1, a form of Mitoferrin 1. Representative PGSK (fluorescent reporter of iron) quenching curves upon addition of iron to TMfrn1 proteoliposome. Red traces are increasing concentrations of iron. Black dotted traces show protein-free liposomes.
Mfrn1 and Mfrn2 were discovered more than 10 years ago. However, the proposed iron transport activity of either transporter has not yet been demonstrated using an in vitro functional reconstitution assay. Also no report about their interaction with iron or other related metal ions exists, most likely because heterologous overexpression and purification of mitoferrins have not yet been reported in the literature. If mitoferrins do indeed transport iron, it is important to know how selective they are for iron over other similar metal ions, what residues in mitoferrin-1 and -2 are important for their function, and whether they even bind iron. Such fundamental questions about mitoferrin function have, to date, not been addressed.
We have now carried out heterologous purification and in vitro functional reconstitution and mutational dissection of a vertebrate Mfrn1 (Figure 2c). This is the first demonstration that Mfrn1 can indeed transport iron. We showed that Mfrn1 is a promiscuous metal ion transporter in that it also transports other first-row transition metal ions. Through mutagenesis, we discovered candidate residues that are important for metal ion transport by Mfrn1 and those that could be involved in forming metal-ion binding sites during transport.
Our studies provide the first biochemical insights into Mfrn function and form the starting point for future high-resolution structural studies of Mfrn function. Our transport assay and the purification strategy will lead to more detailed biochemical and biophysical experiments into the mechanistic basis of iron transport by Mfrn1 and Mfrn2. Our in vitro proteoliposome-reconstituted iron and copper transport assay is likely to be used for studying other iron and copper transporters and, considering the importance of iron and copper transport in biology, these assays will be important tools for biochemical dissection of the transporters of these metal ions.
Molecular mechanism of Porcupine, an integral membrane enzyme of the MBOAT family, which catalyzes Wnt lipidation
In metazoans, Wnt proteins regulate a large number of processes, including cellular growth, differentiation, and tissue homeostasis through the highly conserved Wnt signaling pathway. Porcupine is an endoplasmic reticulum (ER)–resident integral membrane enzyme that catalyzes post-translational modification of Wnts with palmitoleic acid, an unsaturated lipid. This unique form of lipidation with palmitoleic acid is a vital step in the biogenesis and secretion of Wnt, and Porcupine inhibitors are currently in clinical trials for cancer treatment. However, Porcupine-mediated Wnt lipidation has not been reconstituted in vitro with purified enzyme. We recently reported the first successful purification of human Porcupine and confirmed, through in vitro reconstitution with the purified enzyme, that Porcupine is necessary and sufficient for Wnt acylation. By systematically examining a series of substrate variants, we showed that Porcupine intimately recognizes the local structure of Wnt around the site of acylation. Our in vitro assay enabled us to examine the activity of Porcupine with a range of fatty acyl–CoAs with varying length and unsaturation. The selectivity of human Porcupine across a spectrum of fatty acyl–CoAs suggested that the kink in the unsaturated acyl chain is a key determinant in Porcupine-mediated catalysis. We also showed that two putative Porcupine inhibitors, which were discovered with cell-based assays, indeed target human Porcupine. Together, these results provide several, high-resolution biochemical insights into the mechanism of Porcupine-mediated Wnt acylation and pave the way for further detailed biochemical and structural studies.
Click image to view.
Figure 3. Structure of DHHC palmitoyltransferases
a. The structure of human DHHC20 and a catalytically inactive mutant of zebrafish DHHC15 shown in ribbon trace. The transmembrane domain (TM) is shown in green, the DHHC–containing cysteine-rich domain in blue and the C-terminal domain in orange. The grey spheres indicate Zn2+ ions. These are both Golgi-resident enzymes and thus the top side faces the Golgi lumen and the active site the cytoplasm.
b. Active site of human DHHC20 showing the catalytic triad containing the active-site cysteine.
c. Structure of human DHHC20 irreversibly modified with 2-bromopalmitate, which results in the active-site cysteine linking to the alpha-carbon of palmitic acid. The acyl group of palmitic acid is shown in stick rendition. Also shown are two residues towards the top of the tapering cavity to where the palmitate binds.
d. The acyl chain length selectivity of wild-type (WT) human DHHC20. Mutation of tyrosine181 to alanine (Y181A) expands the cavity and shifts the acyl selectivity to the longer side. On the other hand, mutation of serine29 to phenylalanine (S29F) contracts the cavity and thus shifts the acyl selectivity to the shorter side.
Additional Funding
- NIH Intramural AIDS Targeted Antiviral Program (IATAP) award to Anirban Banerjee (2018)
- NICHD DIR Director's Investigator Award
Publications
- Rana MS, Kumar P, Lee CJ, Verardi R, Rajashankar KR, Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 2018;359:6372.
- Rana M, Lee C, Banerjee A. The molecular mechanism of DHHC palmitoyltransferases. Biochem Soc Transactions 2018;BST20180429.
- Christenson ET, Gallegos AS, Banerjee A. In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J Biol Chem 2018;293:3819-3828.
Collaborators
- Rodolfo Ghirlando, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- James Inglese, PhD, Assay Development & Screening Technology Laboratory, NCATS, Bethesda, MD
- Kanagalaghatta R. Rajashankar, PhD, Northeastern-Collaborative Access Team, Argonne National Laboratory and Cornell University, Lemont, IL
- Lei Shi, PhD, Computational Chemistry and Molecular Biophysics Unit, NIDA, Baltimore, MD
Contact
For more information, email anirban.banerjee@nih.gov or visit http://banerjee.nichd.nih.gov.


