Mechanisms of Nuclear Genome Organization and Maintenance
- Mary Dasso, PhD, Head, Section on Cell Cycle Regulation
- Alexei Arnaoutov, PhD, Staff Scientist
- Hang Noh Lee, PhD, Research Fellow
- Maia Ouspenskaia, DVM, Biologist
- Vasilisa Aksenova, PhD, Visiting Fellow
- Shane Chen, PhD, Postdoctoral Intramural Research Training Award Fellow
- Karen M. Plevock Haase, PhD, Postdoctoral Intramural Research Training Award Fellow
- Saroj Regmi, PhD, Postdoctoral Intramural Research Training Award Fellow
- Carlos Echeverria, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Ross T. Kaufhold, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Elizabeth Turcotte, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Ka Chun Yau, BS, Predoctoral Visiting Fellow
We are interested in mechanisms of genome maintenance and organization. During interphase, chromosomes are surrounded by the nuclear envelope (NE), separating the nuclear and cytoplasmic compartments of the cell. Exchange of all molecules between the nucleus and the rest of the cell occurs through nuclear pore complexes (NPCs; Figure 1), which are embedded in the NE. NPCs are not only conduits for nucleo-cytoplasmic trafficking, they also promote many aspects of interphase nuclear function, including gene expression and heterochromatin organization. As mammalian cells divide, the NE breaks down and chromosomes condense to allow their partitioning into daughter cells. Remarkably, NPC proteins (nucleoporins) and other components of the nuclear transport machinery have distinct and important mitotic roles that assure the accurate segregation of chromosomes.
Our focus is to elucidate the role of nucleoporins, NPC–associated proteins (e.g., the SUMO pathway, spindle checkpoint proteins), and other components of the nuclear transport machinery (e.g., the Ran pathway) throughout the cell cycle. We are interested both in how they maintain nuclear organization during interphase and how they promote chromosome segregation. To address both aspects, we adapted CRISPR–based degron strategies to the study of these proteins; namely, we tagged the proteins with AID (auxin-induced degrons; a degron is a short portion of a protein important for protein degradation rates), which cause them to be specifically destroyed upon the addition of auxin, a plant hormone. The ultimate goals of our studies are to understand how these pathways enable correct genome organization and accurate chromosome segregation, as well as to discover how their functions are coordinated with each other and with other aspects of cell physiology.
Click image to view.
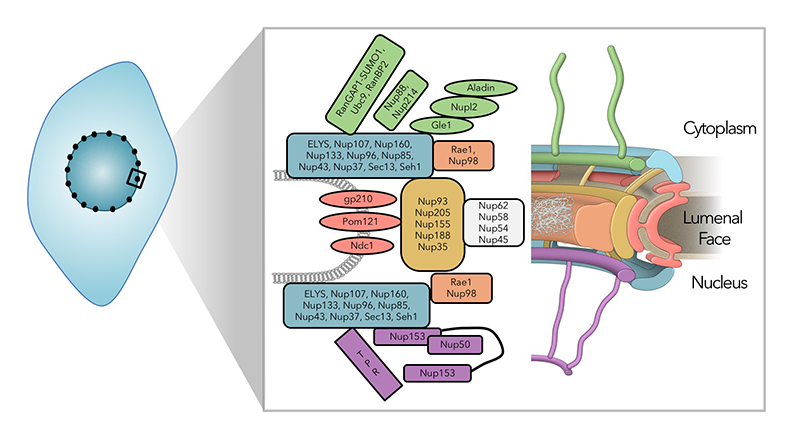
Figure 1. The nuclear pore complex is a large, multi-functional structure.
Schematic representation of the nuclear pore complex (NPC). NPCs are embedded in the nuclear envelopes of all eukaryotic cells, where they are not only conduits for nucleocytoplasmic trafficking, but they also promote many aspects of interphase nuclear function, including gene expression and heterochromatin organization. Around thirty proteins have been identified as nucleoporins through proteomic analysis of NPCs (left), whose corresponding distribution is indicated by color in the structural representation (right).
Targeting nucleo-cytoplasmic transport proteins for selective degradation
NPCs consist of multiple copies of roughly 30 proteins (nucleoporins), which form a series of stable sub-complexes. Many nucleoporins play additional roles during mitosis, and some sub-complexes localize to kinetochores, where they facilitate chromosome segregation. Understanding the activities of individual nucleoporins in each of these contexts has been complicated by their multi-faceted nature: for example, RNAi (RNA interference)–mediated depletion of nucleoporins in mammalian cells causes phenotypic defects, including altered gene expression and abnormal mitotic progression. However, the abundance and unusual stability of these proteins requires an extended interval for their depletion, so that many phenotypes could be indirect consequences of disrupted nuclear trafficking as the number and quality of NPCs decline.
To better address the role of individual nucleoporins, we adapted AID strategies for selective and rapid degradation of individual proteins (Figure 2). We are using CRISPR/Cas9 to construct cell lines in which sequences encoding AIDs are inserted into both alleles of targeted genes of human tissue-culture cells that stably express the Transport Inhibitor Response 1 (TIR1) protein. TIR1 acts as a subunit of the SCF ubiquitin ligase complex, so that the AID–tagged fusion proteins undergo rapid, selective degradation upon addition of the plant hormone auxin. In general, we also add a fluorescent tag to the targeted proteins, allowing the degradation to be monitored visually as well as biochemically.
We have been successful in developing lines that allow conditional depletions of nucleoporins associated with the cytoplasmic and nuclear faces of the NPC, as well as nucleoporins that reside within the central domain of the NPC. We are examining the function of these nucleoporins in interphase and mitosis, as discussed below. We are continuing our efforts to make lines for conditional depletion of all remaining nucleoporins, as well as collaborating extensively with groups who have developed approaches to address other issues of nucleoporin function.
Click image to view.
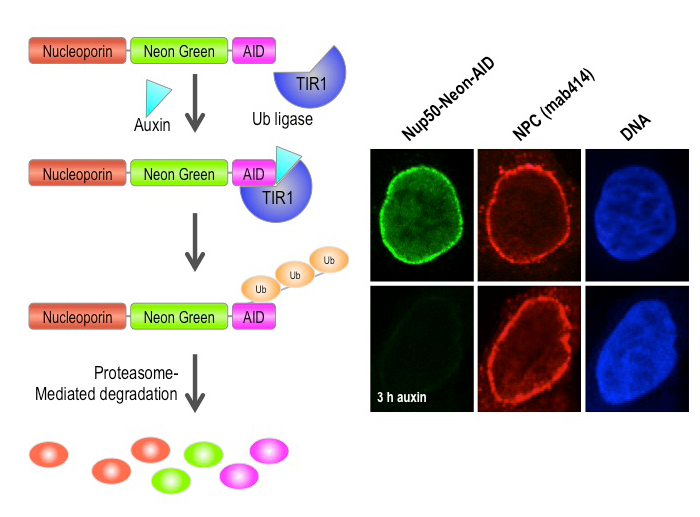
Figure 2. Auxin-induced degradation of AID–tagged nucleoporins
Cells expressing the TIR1 protein recognize proteins tagged with auxin-induced degron (AID) domains upon the addition of the plant hormone auxin. This leads to their rapid conjugation with ubiquitin (Ub) and destruction. We took advantage of this system by homozygously targeting endogenous nucleoporin genes with the AID tag and a fluorescent marker (neon green) in TIR1–expressing DLD1 cells. As shown on the right for the nucleoporin Nup50, we observe rapid and uniform degradation after auxin addition (left panels). Note that the nuclear pore is not generally disrupted, as indicated by staining with an antibody that recognizes a family of nucleoporins (mab414, middle panels).
The role of nucleoporins in interphase nuclear organization and function
Besides nuclear-cytoplasmic transport, NPCs are implicated in the maintenance of nuclear architecture, the organization of interphase chromatin, mRNA export, and transcription regulation. The involvement of the NPC in these various processes offers a rich variety of possible mechanisms for biological regulation via nucleoporins and for coordination among cellular functions. However, for technical reasons these possibilities remain largely unexplored because the abundance and stability of nucleoporins makes them difficult to analyze using standard methods, such as RNAi. Moreover, extended depletion times are associated with the loss of non-targeted nucleoporins and with defects in NPC assembly, leading to further complications in the interpretation of resultant phenotypes. The rapid degradation of nucleoporins using AID–tagged cell lines has helped circumvent some of these issues, and we are currently using them to examine three major aspects of interphase nuclear structure and function.
First, we are examining the role of individual nucleoporins within the NPC structure, testing previously described requirements for individual nucleoporins in the stable assembly of others. We find that the results are distinct depending upon whether we examine the stability of exiting NPC structures or the post-mitotic re-assembly of NPCs. Second, we are examining the role of individual nucleoporins' different nuclear trafficking pathways. We are currently assaying their contributions to nuclear protein import, protein export, and RNA export. We find distinct sensitivities of these pathways to the loss of individual nucleoporins, supporting the idea that the transport pathways have distinct NPC structural requirements. Third, we are examining gene expression by RNA-seq (RNA sequencing) with and without auxin, to observe the consequences of acute and prolonged nucleoporin depletion. Our results suggest that different nucleoporins have distinct roles in gene expression.
Defining which individual nucleoporins contribute to each of these processes will allow better design of future experiments to examine the role of those selected proteins in the regulation particularly of cellular pathways.
Mitotic roles of nuclear pore complex proteins
We have a long-standing interest in the process of chromosome segregation. Defects in chromosome segregation lead to aneuploidy, a condition in which cells possess an abnormal number of chromosomes. Several common birth defects, such as Down's syndrome, result from aneuploidy arising during meiotic cell divisions, and aneuploidy arising from mitotic divisions is a hallmark of many types of solid tumors.
Kinetochores are proteinaceous structures that assemble at the centromere of each sister chromatid during mitosis and serve as sites of spindle microtubule attachment. The relationship between NPCs and mitotic kinetochores is surprisingly intimate but poorly understood. During interphase, many kinetochore proteins stably bind to NPCs (e.g., Mad1, Mad2, Mps1). After mitotic NPC disassembly in mammalian cells, many nucleoporins associate with mitotic spindles and kinetochores, where they play important roles in chromosome segregation.
As discussed above, the multifunctional nature of nucleoporins in controlling in interphase nuclear organization, and nuclear trafficking made it difficult to precisely study their mitotic role using previously available techniques, such as RNAi–based depletion strategies, without disrupting other cell functions. The construction of AID–tagged cell lines allows us to selectively degrade nucleoporins at or near the point of mitotic entry, so that we can circumvent the requirement for these proteins during interphase. We are currently analyzing the roles of individual nucleoporins and NPC–associated protein in kinetochore function, mitotic progression, and spindle assembly.
Mitotic regulation of the Ran GTPase
Ran is a Ras–family GTPase that plays critical roles in many cellular processes, including nucleo-cytoplasmic transport, nuclear envelope assembly, and mitotic spindle assembly. Ran alternates between GDP– and GTP–bound forms. In interphase cells, GTP–bound Ran (Ran–GTP) is the major form in the nucleus, while GDP–bound Ran (Ran-GDP) is the predominant form in the cytoplasm. The asymmetrical distribution of Ran–GTP and Ran-GDP drives cargo transport between the nucleus and cytoplasm through karyopherins, a family of nuclear transport carrier proteins that bind to Ran–GTP. During mitosis, after nuclear envelope breakdown, Ran–GTP is concentrated in the region close to mitotic chromatin, while Ran-GDP is the major form distal to chromatin. The Ran–GTP gradient guides mitotic spindle assembly by releasing spindle assembly factors (SAFs) from karyopherins based on local Ran-GTP concentrations. In cells, the conversion of Ran–GDP to Ran–GTP is catalyzed by a Ran–specific guanine exchange factor called RCC1 (regulator of chromosome condensation 1) in vertebrates. The capacity of RCC1 to bind to chromatin establishes the asymmetrical distribution of Ran–GTP in interphase as well as the chromatin-centered Ran–GTP gradient in mitosis. Interestingly, RCC1’s association with chromatin is not static during the cell cycle and, in vertebrate systems, is regulated in a particularly dramatic fashion during anaphase. The regulation has not been correlated with post-translational modifications of RCC1, and the underlying molecular mechanism has not been reported.
RanBP1 is a highly conserved Ran–GTP–binding protein, which acts as co-activator of RanGAP1 (Ran GTPase–Activating Protein 1) and can form a heterotrimeric complex with Ran and RCC1 in vitro. We found that RCC1 not associated with chromosomes during mitosis is sequestered and inhibited in RCC1/Ran/RanBP1 heterotrimeric complexes and that the sequestration is crucial for normal mitotic spindle assembly. In addition, RanBP1 complex formation competes with chromatin binding to regulate the distribution of RCC1 between the chromatin-associated and soluble fractions. Moreover, we identified a cell cycle–dependent phosphorylation on RanBP1 that modulates RCC1/Ran/RanBP1 heterotrimeric complex assembly and releases RCC1 to bind to chromatin; the phosphorylation is directly responsible for controlling RCC1 dynamics during anaphase. Together, our findings demonstrate novel roles of RanBP1 in spindle assembly and RCC1 regulation in mitosis. We are currently extending these findings to determine whether RanBP1 plays an analogous role in mammalian cells during mitosis.
SUMO–family small ubiquitin-like modifiers in higher eukaryotes
Click image to view.
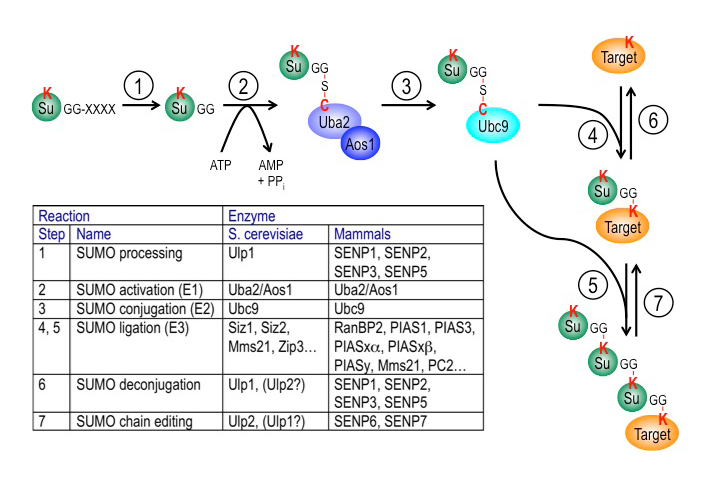
Figure 3. The SUMO pathway
SUMO proteins are post-translationally processed (step 1). Processed SUMO polypeptides possess a C-terminal diglycine motif, which is activated to form an ATP–dependent thioester linkage with the SUMO E1 enzyme, the Aos1/Uba2 heterodimer (step 2). The activated SUMO is transferred to thioester linkage on a conserved cysteine of the SUMO E2 enzyme Ubc9 (step 3). Finally, the activated SUMO becomes covalently linked through an isopeptide bond to lysine residues within cellular target proteins, a reaction that is typically promoted by SUMO ligases (E3 enzymes) acting in conjunction with Ubc9 (step 4). For some substrates, additional SUMOs can be added to form SUMO chains that can act as a signal for proteolytic degradation (step 5). Both mono-SUMOylation (step 6) and poly-SUMOylation (step 7) can be reversed by a family of SUMO–specific proteases that are also major catalysts of post-translational SUMO processing, called Ulps (Ubiquitin-like protein proteases) in yeast and SENPs (Sentrin-specific protease) in vertebrates. The inserted table provides the names of proteins involved in each of these steps in budding yeast and human cells. Note that many of these enzymes (Ubc9, RanBP2, Ulp1, SENP1, SENP2) associate with the nuclear pore complex.
SUMOs are ubiquitin-like proteins (Ubls) that become conjugated to substrates through a pathway that is biochemically similar to ubiquitination (Figure 3). SUMOylation is involved in many cellular processes, including DNA metabolism, gene expression, and cell-cycle progression. Vertebrate cells express three major SUMO paralogs (SUMO-1–3): mature SUMO-2 and SUMO-3 are 95% identical, while SUMO-1 is 45% identical to SUMO-2 or SUMO-3 (where they are functionally indistinguishable, we collectively call SUMO-2 and SUMO-3 SUMO-2/3). Like ubiquitin, SUMO-2/3 can be assembled into polymeric chains through the sequential conjugation of SUMOs to each other. Many SUMOylation substrates have been identified. SUMOylation promotes a variety of fates for individual targets, dependent upon the protein itself, the conjugated paralog, and whether the conjugated species contains a single SUMO or SUMO chains.
SUMOylation is dynamic owing to rapid turnover of conjugated species by SUMO proteases. Both post-translational processing of SUMO polypeptides and deSUMOylation are mediated by the same family of proteases, which play a pivotal role in determining the spectrum of SUMOylated species. This group of proteases is called Ubl–specific proteases (Ulp) in yeast and Sentrin-specific proteases (SENP) in vertebrates. There are two yeast Ulps (Ulp1p and Ulp2p/Smt4p) and six mammalian SENPs (SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7). SENP1, SENP2, SENP3, and SENP5 form a Ulp1p–related sub-family, while SENP6 and SENP7 are more closely related to Ulp2p. Yeast Ulps have important roles in mitotic progression and chromosome segregation. We defined the enzymatic specificity of the vertebrate SENP proteins and analyzed their key biological roles.
Ulp1p localizes to NPCs, is encoded by an essential gene, and is important for SUMO processing, nucleocytoplasmic trafficking, and late steps in the ribosome biogenesis pathway. Humans possess two NPC–associated SENPs: SENP1 and SENP2. While SENP2 is dispensable for cell division, mammalian SENP1 was recently shown to play an essential role in mitotic progression. We are currently analyzing AID–tagged alleles of both SENP1 and SENP2 to assess their roles in both interphase (nuclear trafficking and gene expression) and during mitosis (kinetochore function and mitotic progression), as well as their dependence upon individual nucleoporins for their targeting to the interphase NPC.
The role of the IRBIT protein in tissue homeostasis
We recently reported a conserved role for the IRBIT protein (IP3-receptor-binding protein released with inositol 1,4,5-trisphosphate) in inhibiting ribonucleotide reductase (RNR), an enzyme that produces deoxynucleotide triphosphates (dNTPs) within the cell for DNA synthesis. We further found that mammalian tissue-culture cells show altered cell-cycle progression and potentially disrupted genome stability in the absence of IRBIT. In the same report, we showed that this mechanism is conserved between humans and flies (Drosophila melanogaster). Therefore, in collaboration with Mihaela Serpe and Brian Oliver, we chose to use flies as a model organism to understand the role of this mechanism in development and tissue homeostasis.
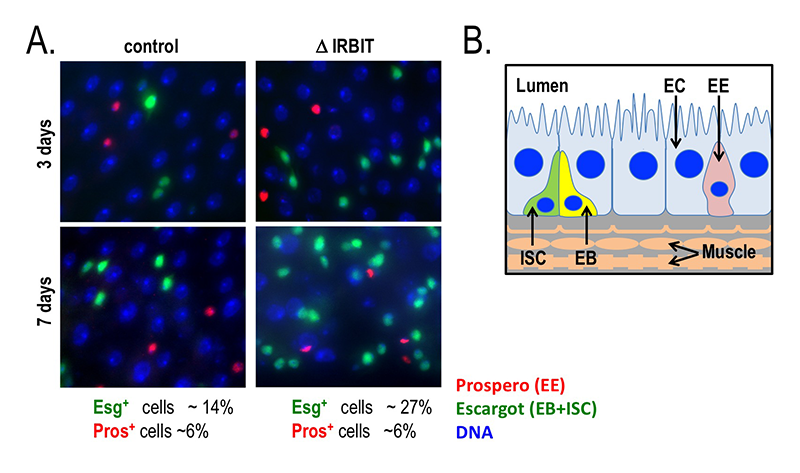
In situ hybridization shows IRBIT expression in regions destined to become the midgut during embryogenesis, and IRBIT is highly expressed in the adult midgut. The Drosophila midgut has a tubular structure and is surrounded by visceral muscles. The adult midgut possesses a monolayered epithelium that is composed of four distinct cell types (Figure 4B): intestinal stem cells (ISCs), undifferentiated progenitor cells called enteroblasts (EBs), specialized absorptive enterocytes (ECs), and secretory enteroendocrine cells (EEs). The midgut is maintained through division of ISCs, giving rise to EBs, which in turn differentiate into EEs. Nutrients are absorbed from the lumen of the gut, which also contains a complex microbiota; the midgut acts both as a niche for commensal microbes and as the first line of defense for against microbial pathogens. Like the intestine of vertebrates, the epithelium of the midgut has a remarkable regenerative capacity, which has been extensively exploited for the study of stem cell–driven tissue self-renewal, as well as tissue homeostasis during aging.
We examined IRBIT’s potential role in the midgut by generating an IRBIT null fly (IRBIT–/–) (Figure 4A). The midguts of one-day-old wild-type and IRBIT–/– flies were essentially indistinguishable at the tissue-architecture level. However, we observed a rapid loss of tissue homeostasis in the IRBIT–/– flies, with a progressive increase in relative numbers of undifferentiated enteroblast progenitor cells and tissue dysplasia. IRBIT–/– flies also show fewer cell-cell contacts when stained for junctional proteins in the posterior midgut epithelium and altered gene expression patterns reminiscent of changes associate with inflammation and aging. The phenotypes are fully rescued through expression of full-length IRBIT, and further experiments suggested that altered dNTP pools likely contribute to the IRBIT–/– phenotypes.
Together, our data suggest that IRBIT plays a key role in tissue homeostasis in the fly midgut by antagonizing RNR in nucleotide metabolism. We are currently examining the interplay of IRBIT with inflammatory pathways and microbiota changes that are characteristic features of aging in flies.
Click image to view.
Figure 4. Loss of IRBIT disrupts tissue homeostasis in the Drosophila midgut.
A. Epithelia from control (left) and IRBIT–/– (right) flies at three (top) or seven (bottom) days after eclosure. The guts were stained with DNA dye Hoechst 33258 (blue), and antibodies against Prospero (EE cells) and Escargot (EB and ISC cells). We observed a rapid and progressive increase in the fraction of Escargot-positive cells in the IRBIT–/– flies over time. In conjunction with additional experiments, the accumulation is indicative of accumulation of undifferentiated enteroblast progenitor cells.
B. Schematic of epithelium within the Drosophila midgut. EC: enterocyte; ISC: intestinal stem cell; EB: enteroblast; EE: enteroendocrine cell.
Additional Funding
- NICHD DIR Director’s Investigator Award
- Developing Talent Scholar Award (Carlos Echeverria)
Publications
- Zhang MS, Furuta M, Arnaoutov A, Dasso M. RCC1 regulates inner centromeric composition in a Ran-independent fashion. Cell Cycle 2018;17(6):739-748.
- Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, Wen X, Parvathaneni S, Kazandjian D, Jenkins LM, Tang W, Elloumi F, Martindale JL, Huarte M, Zhu Y, Robles AI, Frier SM, Rigo F, Cam M, Ambs S, Sharma S, Harris CC, Dasso M, Prasanth KV, Lal A. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep 2017;20(10):2408-2423.
- Chaudhary R, Gryder B, Woods WS, Subramanian M, Jones MF, Li XL, Jenkins LM, Shabalina SA, Mo M, Dasso M, Yang Y, Wakefield LM, Zhu Y, Frier SM, Moriarity BS, Prasanth KV, Perez-Pinera P, Lal A. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife 2017;6:e23244.
- Dasso M, Fontoura BM. Gating immunity and death at the nuclear pore complex. Cell 2016;166:1364-1366.
- Markossian S, Arnaoutov A, Saba NS, Larionov V, Dasso M. Quantitative assessment of chromosome instability induced through chemical disruption of mitotic progression. Cell Cycle 2016;15:1706-1714.
Collaborators
- Job Dekker, PhD, University of Massachusetts Medical School, Worcester, MA
- Beatriz Fontoura, PhD, University of Texas Southwestern Medical Center, Dallas, TX
- Ashish Lal, PhD, Genetics Branch, Center for Cancer Research, NCI, Bethesda, MD
- Song-Tao Liu, PhD, The University of Toledo, Toledo, OH
- Brian C. Oliver, PhD, Laboratory of Cellular and Developmental Biology, NIDDK, Bethesda, MD
- Thomas U. Schwartz, PhD, Massachusetts Institute of Technology, Cambridge, MA
- Mihaela Serpe, PhD, Section on Cellular Communication, NICHD, Bethesda, MD
- Weidong Yang, PhD, Temple University, Philadelphia, PA
Contact
For more information, email mdasso@helix.nih.gov or visit http://sccr.nichd.nih.gov.


