Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Peter Eriksson, PhD, Staff Scientist
- Razvan V. Chereji, PhD, Research Fellow
- Josefina Ocampo, PhD, Visiting Fellow
- Christopher Coey, PhD, Intramural Research Training Award Fellow
- Allison F. Dennis, BS, Graduate Student
- LauraAnn Schmidberger, BS, Postbaccalaureate Fellow
Our main objective is to understand how genes are activated for transcription in the context of chromatin structure. Chromatin is generally repressive in nature, but its structure is manipulated by cells in a regulated way to determine which genes are transcriptionally active and which remain repressed in a given cell type. Chromatin is not just a packaging system for DNA in eukaryotic cells but also participates in gene regulation, which involves either attenuation of the inherently repressive properties of nucleosomes to facilitate gene expression, or enhancement of those properties to ensure complete repression, events that are choreographed by DNA sequence–specific transcription factors (activators and repressors) and chromatin-remodeling complexes. The complexes include histone- or DNA–modifying enzymes that implement the "epigenetic code", and ATP–dependent remodeling machines that move or displace nucleosomes.
The structural subunit of chromatin is the nucleosome core, which contains about 147 bp of DNA wrapped approximately 1.7 times around a central histone octamer. The octamer is composed of two molecules each of the four core histones (H2A, H2B, H3, and H4). Generally, nucleosomes are regularly spaced along the DNA, like beads on a string. At physiological salt concentrations, the beads-on-a-string structure folds spontaneously to form a fiber about 30 nm wide, assisted by the linker histone (H1), which binds to the nucleosome core and to the linker DNA. Thus, collectively, the histones determine DNA accessibility.
Gene activation involves the recruitment of a set of factors to a promoter in response to appropriate signals, ultimately resulting in the formation of an initiation complex by RNA polymerase II (Pol II) and transcription. Nucleosomes are compact structures capable of blocking transcription at every step. To circumvent and regulate the chromatin block, eukaryotic cells possess dedicated enzymes, including ATP–dependent chromatin-remodeling machines, histone-modifying complexes, and histone chaperones. The remodeling machines (e.g., the SWI/SNF, RSC, CHD, and ISWI complexes) use ATP to move nucleosomes along or off DNA or to substitute histone H2A in a nucleosome with the H2A.Z variant (e.g., the SWR complex). The histone-modifying complexes contain enzymes that modify the histones post-translationally to alter their DNA–binding properties and to mark them for recognition by other complexes, which have activating or repressive roles (the "histone code" hypothesis). Histone-modifying enzymes include histone acetylases (HATs), deacetylases (HDACs), methylases, and kinases. Histone chaperones (e.g., Asf1 and the CAF-1 complex) mediate the histone transfer reactions that occur during transcription and DNA replication. These enzymes, together with DNA–methylating and de-methylating enzymes, are central to epigenetics.
Aberrant gene regulation is the basis of many disease states, and many human diseases have been linked to chromatin-remodeling enzymes and epigenetic modifications. For example, mutations in the hSNF5 subunit of the SWI/SNF complex are strongly linked to pediatric rhabdoid tumors. The CHD class of ATP–dependent remodelers has also been linked to cancer and to autism. Cancer therapies and drugs aimed at epigenetic targets are being tested. Recent studies have revealed a correlation between a linker histone variant and tumor heterogeneity. A full understanding of the functions of chromatin structure, enzymes, and modifications is therefore vital.
We are exploiting and developing high-throughput technologies to obtain genome-wide maps of nucleosomes, chromatin-remodeling complexes and RNA polymerase II, etc. We have made major progress in our genomics studies, primarily using budding yeast (Saccharomyces cerevisiae) as a model organism. Budding yeast possesses homologs of most chromatin-modifying enzymes found in mice and humans. It provides a simple, experimentally tractable model for understanding the functions and mechanisms of action of these enzymes. Such studies are facilitating the design and execution of the more complex and difficult experiments that are necessary to understand the roles of chromatin-remodeling enzymes in higher organisms.
The current objectives of our yeast studies are to: (1) determine the roles of various chromatin remodeling complexes (RSC, SWI/SNF, ISW1, ISW2, and CHD1) in chromatin organization and gene expression, which are important because genes encoding homologous subunits of some of these enzymes are often mutated in various cancers; and (2) understand the nature and role of stable non-histone "barrier" complexes bound at active promoters. Through collaborations, we are now extending our studies on chromatin remodeling from yeast to mouse.
Roles of the SWI/SNF and RSC chromatin-remodeling complexes in gene activation by the Gcn4 transcription factor
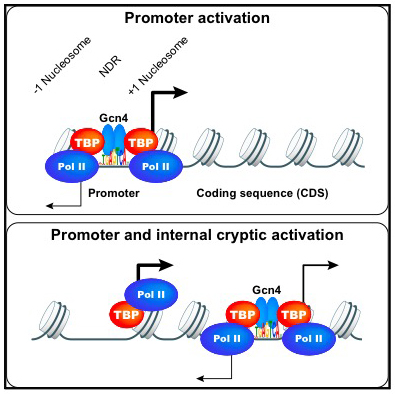
We continued our yeast studies in collaboration with Alan Hinnebusch's lab. In our first collaborative project [Reference 1], we investigated transcriptional activation by the well studied yeast transcription factor Gcn4 in vivo. Gcn4 production is under translational control and is induced by amino acid starvation. Gcn4 then binds to and activates transcription from specific promoters for genes encoding proteins required for amino acid biosynthesis. We expected that specific Gcn4 binding would be limited to promoter sites. Surprisingly, our ChIP-seq data for Gcn4 showed that the majority of sequence-specific DNA binding sites are not in gene promoters, but inside coding regions. Moreover, Gcn4–bound sites within coding regions are not associated with the nucleosome-depleted regions (NDRs) typically observed at promoter sites. Nevertheless, we found that many of the internal Gcn4 sites activate cryptic internal promoters and, in some cases, neighboring promoters, in a manner reminiscent of the transcription enhancers typically found in higher eukaryotes (Figure 1).
Click image to view.
Figure 1. Two modes of gene activation by the yeast Gcn4 transcription activator
1. Conventional promoter binding by Gcn4 results in transcription by Pol II.
2. Internal bidirectional cryptic promoter activation by Gcn4. Adapted from Reference 1.
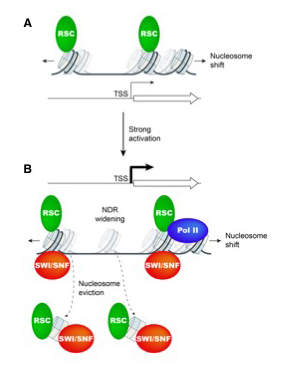
In a second collaborative project with the Hinnebusch lab and with Chhabi Govind's lab, we explored the roles of the related RSC and SWI/SNF remodelers in gene activation [Reference 2]. We had previously shown that the RSC nucleosome remodeling complex functions throughout the yeast genome to set the positions of the nucleosomes flanking promoters (the +1 and -1 nucleosomes), thereby determining the widths of the NDRs. The related SWI/SNF complex participates in nucleosome remodeling/eviction and promoter activation at a subset of yeast genes, including those activated by Gcn4, but does not act globally to establish NDRs. By analyzing the large cohort of Gcn4–induced genes in mutants lacking the catalytic subunits of SWI/SNF or RSC, we uncovered cooperation between the two remodelers during transcriptional activation, which together widen the NDR. SWI/SNF functions on a par with RSC at the most highly transcribed, constitutively expressed genes, suggesting general cooperation by these remodelers for maximal transcription (Figure 2). SWI/SNF and RSC occupancies are greatest at the most highly expressed genes, consistent with cooperative functions in nucleosome remodeling and transcriptional activation. At less active promoters, RSC is more important for chromatin remodeling than SWI/SNF.
Click image to view.
Figure 2. Model for cooperation between chromatin remodelers SWI/SNF and RSC in promoter nucleosome disassembly and re-positioning at highly expressed yeast genes
A. At weakly expressed genes, relatively low levels of RSC (green oval) suffice to maintain wild-type NDR formation, primarily by keeping nucleosomes (sectored cylinders) from crossing NDR boundaries. Cylinders with faint shades indicate nucleosome positions in cells depleted of functional RSC.
B. Upon gene activation by Gcn4 or at highly expressed constitutive genes, SWI/SNF (red oval) and RSC are actively recruited and cooperate in evicting and repositioning the −1 nucleosome and the +1 nucleosome to both widen the NDR and keep nucleosomes from assembling there. Cylinders with faint shades indicate nucleosome positions in cells lacking SWI/SNF or depleted of RSC. Pol II may also contribute to eviction of +1 nucleosomes. Adapted from Reference 2.
Major determinants of nucleosome positioning
The compact structure of the nucleosome limits DNA accessibility and inhibits the binding of most sequence-specific proteins. Nucleosomes are not randomly located on the DNA but positioned with respect to the DNA sequence, suggesting models in which critical binding sites are either exposed in the linker, resulting in activation, or buried inside a nucleosome, resulting in repression. The mechanisms determining nucleosome positioning are therefore of paramount importance for understanding gene regulation and other events that occur in chromatin, such as transcription, replication, and repair. In a review [Reference 3], we assessed our current understanding of the major determinants of nucleosome positioning: DNA sequence, non-histone DNA-binding proteins, chromatin-remodeling enzymes, and transcription. We outline the major challenges for the future: elucidating the precise mechanisms of chromatin opening and promoter activation, identifying the complexes that occupy promoters, and understanding the multiscale problem of chromatin fiber organization.
Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo
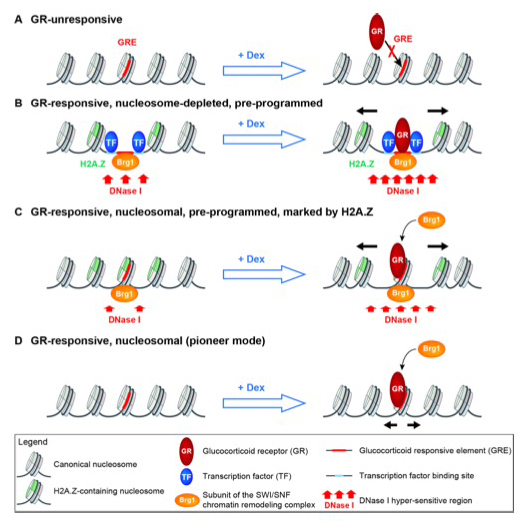
The mechanism by which glucocorticoid hormone controls gene expression is central to an understanding of its role in human metabolism and disease. When the hormone enters a cell, it binds to the glucocorticoid receptor (GR), inducing its translocation from the cytosol to the nucleus, where it binds to glucocorticoid response elements (GREs) to activate or repress target genes. Importantly, GR binds to distinct GREs in different types of cell, resulting in different patterns of gene expression. This observation raises the question as to what distinguishes those sites that are bound from those that are not. Although the answer is unclear, chromatin structure clearly plays a major role. We collaborated with Gordon Hager's lab to study the interaction of the glucocorticoid receptor with enhancer chromatin in mouse adenocarcinoma cells [Reference 4]. We found that enhancers containing GREs occur in one of three major chromatin architectures: they can be nucleosome-depleted, marked by the histone variant H2A.Z in the flanking nucleosomes and associated with the Brg1 SWI/SNF chromatin remodeler; they can be covered by a nucleosome containing H2A.Z and associated with Brg1; or they can be nucleosomal and lack both H2A.Z and Brg1. Hormone-induced GR binding results in nucleosome shifts and increased levels of Brg1 at all three types of GR enhancer. We propose that nucleosome-depleted GR enhancers are created and maintained by hormone-independent transcription factors that recruit Brg1, allowing GR to bind to non-nucleosomal DNA (a "conventional" mode of transcription factor binding). In contrast, at nucleosomal GR enhancers, GR binds like a pioneer factor (i.e., a transcription factor that binds to protein-free and nucleosomal cognate sites with similar affinities) and then recruits Brg1 to remodel the local chromatin structure, promoting downstream events in gene regulation (Figure 3).
Click image to view.
Figure 3. Conventional and pioneer modes of glucocorticoid receptor (GR) binding to responsive enhancers
A. GR does not bind to unresponsive enhancers. They are nucleosomal, inaccessible to DNase I, do not incorporate the histone variant H2A.Z, and lack the Brg1 subunit of the mouse SWI/SNF complex.
B. GR binds to pre-programmed nucleosome-depleted enhancers. The nucleosome-depleted region (NDR) may be created by other transcription factors bound to their cognate sites near the Glucocorticoid Response Element (GRE), in concert with the Brg1 complex, a chromatin remodeler capable of shifting nucleosomes. The NDR is flanked by nucleosomes incorporating H2A.Z and is accessible to DNase I. In response to hormone, GR binds to the GRE in the NDR, and the flanking nucleosomes are shifted aside (black arrows), presumably by Brg1.
C. GR binds to pre-programmed DNase I–accessible nucleosomal enhancers. The GRE is covered by a nucleosome incorporating H2A.Z and associated with Brg1, both contributing to DNase I accessibility. In response to hormone, GR binds to the GRE, recruits more Brg1, and the nucleosome is shifted aside.
D. GR binds to DNase I–insensitive nucleosomal enhancers. The GRE is covered by a nucleosome that lacks H2A.Z and Brg1. In response to hormone, GR binds to the GRE, recruits Brg1, and the nucleosome is shifted. Adapted from Reference 3.
SPT6 interacts with NSD2 and facilitates interferon-induced transcription.
Transcription through chromatin by RNA polymerase II is facilitated by various factors, including the SPT6 histone chaperone. We collaborated with Keiko Ozato's lab to study the role of SPT6 in interferon-induced transcription in mouse cells [Reference 5]. We found that SPT6 recruitment to interferon-induced genes depends on an interaction with the histone H3-K36 methyltransferase NSD2, which boosts transcription of these genes.
Publications
- Rawal Y, Chereji RV, Valabhoju V, Qiu H, Ocampo J, Clark DJ, Hinnebusch AG. Gcn4 binding in coding regions can activate internal and canonical 5' promoters in yeast. Mol Cell 2018;70:297-311.
- Rawal Y, Chereji RV, Qiu H, Ananthakrishnan S, Govind CK, Clark DJ, Hinnebusch AG. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev 2018;32:695-710.
- Chereji RV, Clark DJ. Major determinants of nucleosome positioning. Biophys J 2018;114:2279-2289.
- Johnson TA, Chereji RV, Stavreva DA, Morris SA, Hager GL, Clark DJ. Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res 2018;46:203-214.
- Ouda R, Sarai N, Nehru V, Patel MC, Debrosse M, Bachu M, Chereji RV, Eriksson PR, Clark DJ, Ozato K. SPT6 interacts with NSD2 and facilitates interferon-induced transcription. FEBS Lett 2018;592:1681-1692.
Collaborators
- Harold Burgess, PhD, Section on Behavioral Neurogenetics, NICHD, Bethesda, MD
- Douglas Fields, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Chhabi Govind, PhD, Oakland University, Rochester, MI
- Gordon L. Hager, PhD, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, NCI, Bethesda, MD
- Alan G. Hinnebusch, PhD, Section on Nutrient Control of Gene Expression, NICHD, Bethesda, MD
- Philip R. Lee, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Keiko Ozato, PhD, Section on Molecular Genetics of Immunity, NICHD, Bethesda, MD
Contact
For more information, email clarkda@mail.nih.gov or visit http://clarklab.nichd.nih.gov.


