Cell Cycle Regulation in Oogenesis

- Mary Lilly, PhD, Head, Section on Gamete Development
- Chun-yuan Ting, PhD, Staff Scientist
- Kuikwon Kim, MS, Technician
- Lucia Bettedi, PhD, Visiting Fellow
- Shu Yang, PhD, Visiting Fellow
- Yingbiao Zhang Zhang, PhD, Visiting Fellow
- Christa Ventresca, BS, Postbaccalaureate Fellow
Our long-term goal is to obtain a comprehensive understanding of how metabolic signaling pathways influence oocyte growth, development, and quality. Chromosome mis-segregation during female meiosis is the leading cause of miscarriages and birth defects in humans. Recent evidence suggests that many meiotic errors occur downstream of defects in oocyte growth and/or the hormonal signaling pathways that drive differentiation of the oocyte. Thus, understanding how oocyte development and growth impact meiotic progression is essential to studies in both reproductive biology and medicine. We use the genetically tractable model organism Drosophila melanogaster to examine how meiotic progression is instructed by the developmental and metabolic program of the egg.
In mammals, studies on the early stages of oogenesis face serious technical challenges in that entry into the meiotic cycle, meiotic recombination, and the initiation of the highly-conserved prophase I arrest all occur during embryogenesis. By contrast, in Drosophila these critical events of early oogenesis all take place continuously within the adult female. Easy access to the early stages of oogenesis, coupled with available genetic and molecular genetic tools, makes Drosophila an excellent model for studies on the role of metabolism in oocyte development and maintenance.
The GATOR complex: integrating developmental and metabolic signals in oogenesis
The Target of Rapamycin Complex 1 (TORC1) regulates cell growth and metabolism in response to multiple inputs, including amino-acid availability and intracellular energy status. In the presence of sufficient nutrients and appropriate growth signals, the Ragulator and the Rag GTPases (a complex that regulates lysosomal signaling and trafficking) target TORC1 to lysosomal membranes where TORC1 associates with its activator, the small GTPase Rheb. Once activated, TORC1 is competent to phosphorylate its downstream targets. The Gap Activity Towards Rags (GATOR) complex is an upstream regulator of TORC1 activity.
The GATOR complex consists of two sub-complexes. The GATOR1 complex inhibits TORC1 activity in response to amino-acid starvation. GATOR1 is a trimeric protein complex consisting of the proteins Nprl2, Nprl3, and Iml1. Evidence from yeast and mammals indicates that the components of the GATOR1 complex function as GTPase–activating proteins (GAP) that inhibit TORC1 activity by inactivating the Rag GTPases. Notably, Nprl2 and Iml1 are tumor suppressor genes, while mutations in Iml1, known as DEPDC5 in mammals, are a leading cause of hereditary epilepsy.
The GATOR2 complex comprises five proteins: Seh1, Sec13, Mio, Wdr24, and Wdr59. Our work, as well as that of others, found that the GATOR2 complex activates TORC1 by opposing the TORC1–inhibitory activity of GATOR1. Intriguingly, computational analysis indicates that Mio and Seh1, as well as several other members of the GATOR2 complex, have structural features consistent with coatomer proteins and membrane-tethering complexes. In line with the structural similarity to proteins that influence membrane curvature, we showed that three components of the GATOR2 complex, Mio, Seh1, and Wdr24, localize to the outer surface of lysosomes, the site of TORC1 regulation. However, how GATOR2 inhibits GATOR1 activity, thus allowing for the robust activation of TORC1, remains unknown. Additionally, the role of the GATOR1 and GATOR2 complexes in both the development and physiology of multicellular animals remains poorly defined. Over the last year, we used molecular, genetic, and cell-biological approaches to define the role of the GATOR complex in the regulation of Drosophila oocyte development and physiology.
The GATOR complex regulates an essential response to meiotic double-stranded breaks.
The TORC1 inhibitor GATOR1 controls meiotic entry and early meiotic events in yeast. However, how metabolic pathways influence meiotic progression in metazoans remains poorly understood. In the last year, we determined that the TORC1 regulators GATOR1 and GATOR2 mediate a response to meiotic DNA double-stranded breaks (DSBs) during Drosophila oogenesis. The initiation of homologous recombination through the programmed generation of DNA DSBs is a universal feature of meiosis. DSBs represent a dangerous form of DNA damage that can result in dramatic and permanent changes to the germline genome. To minimize this destructive potential, the generation and repair of meiotic DSBs is tightly controlled in space and time. We showed that meiotic DSBs promote the GATOR1–dependent downregulation of TORC1 activity. These data suggested that low TORC1 activity may be important for the efficient repair of meiotic DSB. In line with this hypothesis, we determined that GATOR1–mutant ovaries, which have high levels of TORC1 activity, exhibit many phenotypes consistent with the misregulation of meiotic DSBs, including an increase in the steady-state number of meiotic DSBs, the retention of meiotic DSBs into later stages of oogenesis, and the hyper-activation of p53, a transcription factor that mediates a highly conserved response to genotoxic stress (Figure 1). Importantly, RNAi depletions of Tsc1 phenocopied the GATOR1 ovarian defects. TSC1 is a component of the potent TORC1 inhibitor Tuberous Sclerosis Complex (TSC), confirming that the misregulation of meiotic DSBs observed in GATOR1–mutant oocytes is attributable to high TORC1 activity rather than to a TORC1–independent function of the GATOR1 complex. Further genetic analysis demonstrated that GATOR1 impacts the repair, rather than the generation, of meiotic DSBs. These data are particularly intriguing in light of similar meiotic defects observed in npr3 mutants in Saccharomyces cerevisiae. Our results raise the possibility that GATOR1–mediated down regulation of TORC1 activity may be a common feature of the early meiotic cycle in many eukaryotes.
Click image to view.
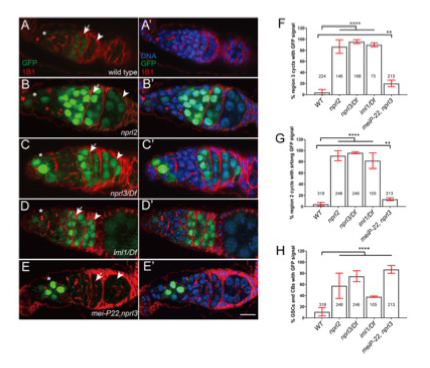
Figure 1. GATOR1 prevents p53 hyperactivation in Drosophila early ovarian cysts.
Ovaries from (A) p53R–GFP, (B) nprl21; p53R–GFP, (C) p53R–GFP;nprl31/Df, (D) p53R–GFP; iml1/Df, and (E) p53R–GFP; mei-P22p22, nprl31 were stained for GFP (green) and 1B1 (red). Germarial regions are defined by 1B1 staining. In wild-type ovaries the p53–GFP reporter is briefly activated in region 2 (indicated by arrow). Note the low level of GFP staining. In contrast, in GATOR1 mutants, p53R–GFP is robustly activated, with strong GFP signal often persisting into germarial region 3 and beyond. Additionally, in GATOR1–mutant germaria, p53R–GFP is frequently activated in germline stem cells (GSC) and daughter cystoblasts (CB). In mei-P22p22, nprl31 double mutant germaria, the hyperactivation of p53R–GFP is rescued in region 2a ovarian cysts. However, p53–GFP activation in GSC and CB is retained in the double mutants (asterisk), indicating that in these cells the activation of p53 is not contingent on the presence of meiotic DSBs. Scale bars, 10μm. (F) Percentage of germaria that sustain p53R–GFP signal in region 3. (G) Percentage of germaria with high p53R–GFP signal in region 2. (H) Percentage of germaria with p53R–GFP expression in GSC and CB. Unpaired T-student test was used to calculate the statistical significance. Error bars represent SD from at least three independent experiments. **P < 0.01, ****P < 0.0001.
GATOR1 prevents the derepression of retrotransposons in response to meiotic double-stranded breaks.
Genotoxic stress has been implicated in the deregulation of retrotransposon expression in several organisms, including Drosophila. In line with these studies, we find that, in GATOR1 mutants, the double-stranded breaks that initiate meiotic recombination trigger the deregulation of retrotransposon expression. Similarly, it was previously shown that p53–mutant females de-repress retrotransposon expression during oogenesis, but as observed in GATOR1 mutants, primarily in the presence of meiotic DSBs. Through epistasis analysis we determined that p53 and GATOR1 act through independent pathways to repress retrotransposon expression in the female germline. Surprisingly, we found that depletions of the potent TORC1 inhibitor TSC in the female germline resulted in little to no increase in retrotransposon expression. The data raise the interesting possibility that GATOR1 regulates retrotransposon expression independently of TORC1 activity. Notably, GATOR1 components, but not TSC components, were recently identified in a high-throughput screen for genes that suppress LINE1 (Long Interspersed Element-1) expression in mammalian tissue culture cells. Taken together, our data indicate that the GATOR1 complex opposes retrotransposon expression during meiosis in a pathway that functions in parallel to p53 in the female germline of Drosophila.
A genetic screen for new regulators of the GATOR complex
We previously demonstrated that mutations in the GATOR2 components mio and seh1 cause constitutive activation of the GATOR1 complex in the female germline, resulting in permanent inhibition of TORC1 activity and a block in oocyte growth and development. Germline RNAi depletions of any of the GATOR1 components in the mio or seh1 mutant backgrounds relieve the permanent TORC1 inhibition and rescue the mio and seh1 ovarian phenotypes (Figure 2). We utilized this epistatic relationship to conduct a high-throughput RNAi–based screen to identify upstream regulators and downstream effectors of the GATOR1 complex during oogenesis. To identify additional genes that, when co-depleted with seh1, rescue the seh1RNAi ovarian phenotypes, we used RNAi lines from the Transgenic RNAi Project (TRiP) that have been optimized for germline expression. We screened 3,472 TRiP collection lines, representing 3,103 target genes, to identify genes that, when co-depleted with seh1, rescue the seh1RNAi ovarian growth phenotype. From the screen, we identified 67 genes, 2.0% of those screened, in which co-expression in the female germline fully suppressed the seh1 ovarian growth deficit. Five genes were identified by two TRiP lines targeting independent sequences. Several lines of evidence indicate that the screen was successful. Importantly, we identified many genes that negatively regulate growth and metabolism, including negative regulators of TORC1/PI3K/insulin signaling, such as pten, Ampk, gigas, mapk (rl), ras and p22AB55 (twins), and pp2A-C (mts), as well as many negative regulators of Hippo, a second signaling pathway that promotes cell proliferation and growth. Gene categories highly enriched in gene-ontology (GO, a comprehensive resource for knowledge regarding the functions of genes and gene products) biological processes include those for: (1) the mTor signaling pathway; (2) autophagy; and (3) the MAPK signaling pathway. Taken together, the findings strongly suggest that the screen provided a sensitized background that successfully identified several pathways regulating germline growth and metabolism. Importantly, our screen identified many genes and pathways not previously associated with growth control during oogenesis. The identified pathways include genes involved in glycolysis, fatty acid metabolism, DNA repair, and stress granule formation, as well as genes of unknown function. The genes will be a rich source for future studies on the metabolic regulation of growth control and meiotic progression during oogenesis.
Click image to view.
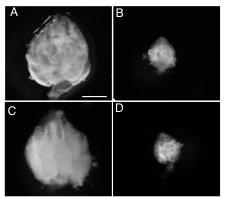
Figure 2. seh1 RNAi depletion and co-depletion phenotypes
Representative images of (A) wild-type (WT) (large ovary), (B) nanos>seh1RNAi (small ovary), (C) nanos>seh1RNAi, nprl2RNAi (large ovary), (D) nanos>seh1RNAi, mCherryRNAi (small ovary). Note that co-depletion of nprl2 rescues the seh1RNAi phenotype, while co-depletion of mCherry fails to rescue the seh1RNAi phenotype.
The GATOR2 component Wdr24 regulates TORC1 activity and lysosome function.
While the GATOR1 complex has been implicated in a wide array of human pathologies, including cancer and hereditary forms of epilepsy, the in vivo relevance in metazoans of the GATOR2 complex remains poorly understood. We defined the in vivo role of the GATOR2 component Wdr24 in Drosophila. Using a combination of genetic, biochemical, and cell-biological techniques, we demonstrated that Wdr24 has both TORC1–dependent and TORC1–independent functions in the regulation of cellular metabolism. Through the characterization of a null allele, we found that Wdr24 is a critical effector of the GATOR2 complex required for the robust activation of TORC1 and cellular growth in a broad array of Drosophila tissues (Figure 3). Additionally, epistasis analysis between wdr24 and genes that encode components of the GATOR1 complex revealed that Wdr24 has a second critical function: the TORC1–independent regulation of lysosome dynamics and autophagic flux. Notably, we found that two additional members of the GATOR2 complex, Mio and Seh1, also play a TORC1–independent role in the regulation of lysosome function. The results represent a surprising and previously unrecognized function of GATOR2 complex components in the regulation of lysosome structure and function. Consistent with our findings in Drosophila, through the characterization of a wdr24–/– knockout HeLa cell line, we determined that Wdr24 promotes lysosome acidification and autophagic flux in mammalian cells. Taken together, our data support a model in which Wdr24 is a key effector of the GATOR2 complex, required for both TORC1 activation and the TORC1–independent regulation of lysosomes. Moreover, our data raise the interesting possibility that the GATOR2 complex regulates GATOR1 through the control of lysosome structure and/or function.
Click image to view.
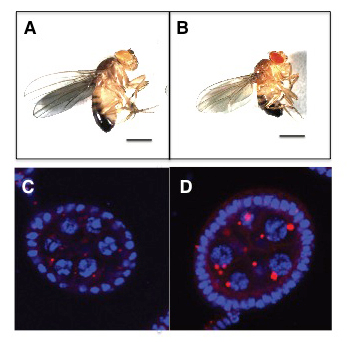
Figure 3. The GATOR2 component Wdr24 regulates growth and autophagy.
Representative images of (A) WT and (B) wdr241–mutant adult males. wdr241 males are notably smaller than WT males. Size bar is 100 μm. Representative images of (C) WT and (D) wdr241 egg chambers from well fed females stained with DAPI to mark DNA (blue) and LysoTracker (red) to mark autolysosomes. Note that wdr241 egg chambers accumulate large numbers of autolysosome-like structures under nutrient-replete conditions.
Publications
- Wei Y, Reveal B, Cai, W, Lilly MA. The GATOR1 complex regulates metabolic homeostasis and the response to nutrient stress in Drosophila. G3 (Bethesda) 2016 6(12):3859-3867.
- Cai W, Wei Y, Jarnik M, Reich J, Lilly MA. The GATOR2 component Wdr24 regulates TORC1 activity and lysosome function. PLoS Genetics 2016 12(5):e1006036.
Collaborators
- Juan Bonifacino, PhD, Section on Intracellular Protein Trafficking, NICHD, Bethesda, MD
- Brian Calvi, PhD, Indiana University, Bloomington, IN
- R. Daniel Camerini-Otero, PhD, Genetics and Biochemistry Branch, NIDDK, Bethesda, MD
- Mary Dasso, PhD, Section on Cell Cycle Regulation, NICHD, Bethesda, MD
- Jan LaRoque, PhD, Georgetown University, Washington, DC
- Yikang Rong, PhD, San Yat-sen University, Guangzho, China
- Erik Snapp, PhD, Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA
Contact
For more information, email mlilly@helix.nih.gov or visit http://cbmp.nichd.nih.gov/uccr.


