The Biological Impact of Transposable Elements

- Henry L. Levin, PhD, Head, Section on Eukaryotic Transposable Elements
- Angela Atwood-Moore, BA, Senior Research Assistant
- Caroline Esnault, PhD, Visiting Fellow
- Feng Li, PhD, Visiting Fellow
- Si Young Lee, PhD, Postdoctoral Fellow
- Zelia Worman, PhD, Postdoctoral Fellow
- Oluwadamilola Bankole, BA, Postbaccalaureate Fellow
- Angelique Ealy, BA, Postbaccalaureate Fellow
- Arianna Lechsinska, BA, Postbaccalaureate Fellow
- Michael Lee, BA, Postbaccalaureate Fellow
- Lauren Tracy, BA, Postbaccalaureate Fellow
- Katie Wendover, BA, Postbaccalaureate Fellow
Inherently mutagenic, the integration of retroviral and retrotransposon DNA is responsible for many pathologies, including malignancy. Given that some chromosomal regions are virtually gene free while others encode genes essential for cellular processes, the position of integration has great significance. Recent studies show clearly that integration occurs into specific types of sequences and that the targeting patterns vary depending on the specific retrovirus or retrotransposon. Currently, there is great interest in such patterns, in part because understanding the mechanisms that position HIV-1 insertions may lead to new antiviral therapies. In addition, retrovirus-based vectors are now being used for gene therapy. Early gene therapy vectors had patterns of integration that activated oncogenes and caused leukemia in patients. It is therefore essential to understand the mechanisms that position such integration. Our current work adopts the methods of high-throughput sequencing to study dense integration patterns of model elements such as the long terminal repeat (LTR) retrotransposon Tf1 of Schizosaccharomyces pombe. This model element allows us to study integration mechanisms using the highly informative techniques of yeast genetics. As an example, we generated an expression technique that tags each integration with a highly specific serial number. With the method, we sequenced 500,000 independent integration events. The improvements we made in sequencing technology are general and allowed us to generate dense profiles of HIV-1 integration. Our analyses of the datasets has greatly improved our understanding of integration and the mechanisms that select insertion sites.
Single nucleotide–specific targeting of the Tf1 retrotransposon promoted by the DNA–binding protein Sap1 of Schizosaccharomyces pombe
Our initial use of deep sequencing revealed that Tf1 integration favors the promoters of RNA polymerase II (RNA pol II)–transcribed genes. In particular, the promoters of stress-response genes are strong targets. As DNA sequencing methods improved, it became possible to map a million integration events of Tf1 within S. pombe. A significant shortcoming of such dense maps of integration is the inability to measure repeated insertions at specific nucleotide positions. This is because we and others discard duplicate sequence reads to avoid PCR–generated distortion. We addressed the problem by including a random eight-nucleotide serial number in the LTR of Tf1. With the method we can count the number of independent insertions at single nucleotide positions. While the serial number system identified specific sequence locations with high integration efficiency, sequence itself did not account for the selection of promoters. We had tested transcription factors known to activate stress-response promoters and found that they do not contribute to the efficiency or position of Tf1 integration. However, a recent study of Switch-activating protein 1 (Sap1), an essential DNA–binding protein in S. pombe, showed that Sap1 binds to genomic positions where Tf1 integration occurs. To determine whether Sap1 plays a role in Tf1 retrotransposition, we studied S. pombe with the temperature-sensitive mutant sap1-1 [Reference 1]. At permissive temperature, Tf1 transposition was reduced ten-fold compared with wild-type sap1+, and the defect was not the result of lower levels of Tf1 proteins or cDNA. The data argue that Sap1 contributes to the integration of Tf1. A mutation that results in 10-fold less integration might be expected to cause off-target integration. Indeed, serial number sequencing of integration in cells with the sap1-1 mutation showed position changes in 10% of the integration events.
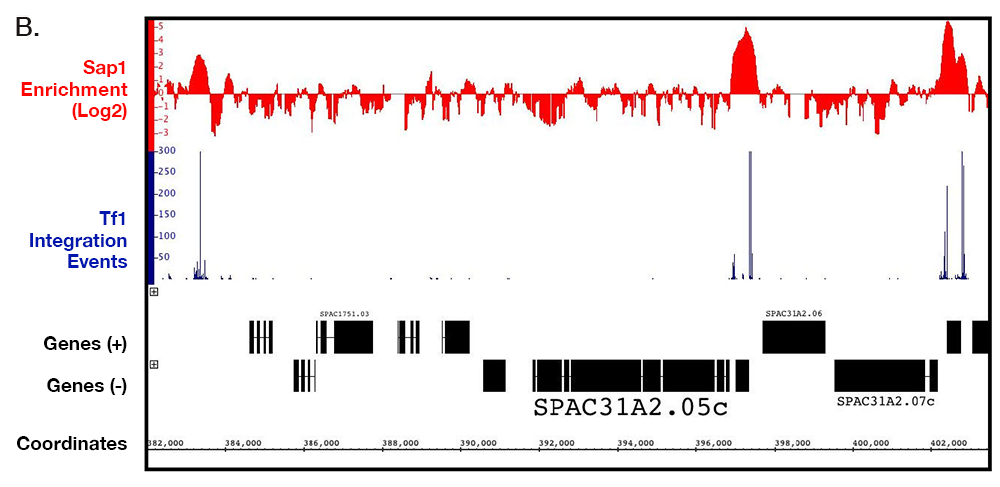
In another approach to determine whether Sap1 contributes to integration, we compared the integration data from the serial number system with previously published maps of Sap1 binding created with ChIP-seq. Analysis of the ChIP-seq data showed that 6.85% of the S. pombe genome was bound by Sap1. Importantly, we found that 73.4% of Tf1 insertions occurred within these Sap1–bound sequences [Reference 1]. An example of this close association can be seen in a segment of chromosome 1 (Figure 1). Another important observation was a strong correlation between levels of integration in intragenic sequences and the amount of Sap1 bound. If Sap1 were directly responsible for positioning Tf1 integration, we would expect integration to take place at specific nucleotide positions relative to the nucleotides bound by Sap1. Using the ChIP-Seq data, we were able to identify a Sap1–binding motif, which closely resembled previously published motifs. We used the FIMO program of the MEME Suite to perform genomic searches, which identified 5,013 locations that matched this motif. The alignment of all these motifs revealed that 82% of all integration events cluster within 1 kb of this motif. Importantly, 43% of all integrations occurred within 50 bp of the motif and they had two dominant positions: 9 bp upstream and 19 bp downstream of the motif. The clustering of inserts at the Sap1 motif would be expected to occur if Sap1 covers its binding site on the DNA and directs integration to either side of the protein. Thus far, we have been unable to detect a direct interaction between Sap1 and Tf1 integrase (IN) with pull-down assays. However, our two-hybrid assays detected a strong Sap1–IN interaction. The two-hybrid result together with the strong alignments of integration with Sap1 motif sequence and the reduction in integration in the sap1-1 mutant argue that Sap1 plays an important role in Tf1 integration.
Click image to view.
Figure 1. Serial number integration data correlates with the position of Sap1 enrichment from ChIP-seq data.
A representative segment of chromosome 1 is shown.
Host factors that promote retrotransposon integration are similar in distantly related eukaryotes.
Retroviruses and LTR retrotransposons have distinct patterns of integration sites. The oncogenic potential of retrovirus-based vectors used in gene therapy depends on the selection of integration sites associated with promoters. The LTR retrotransposon Tf1 of Schizosaccharomyces pombe is studied as a model for oncogenic retroviruses because it integrates into the promoters of stress-response genes. Although INs encoded by retroviruses and LTR retrotransposons are responsible for catalyzing the insertion of cDNA into the host genome, distinct host factors are required for the efficiency and specificity of integration. Our finding that Sap1 is located at positions of integration but does not interact with integrase suggested that other host factors are required for integration. We tested this hypothesis with a genome-wide screen of host factors that promote Tf1 integration. By combining an assay for transposition with a genetic assay that measures cDNA present in the nucleus, we could identify factors that contribute to integration. We used this assay to test a collection of 3,004 S. pombe strains with single-gene deletions [Reference 2]. Using these screens and immunoblot measures of Tf1 proteins, we identified a total of 61 genes that promote integration. The candidate integration factors participate in a range of processes including nuclear transport, transcription, mRNA processing, vesicle transport, chromatin structure, and DNA repair. We tested two candidates, Rhp18 and the NineTeen complex, in two-hybrid assays and found that they interact with Tf1 IN. Surprisingly, several pathways we identified were previously found to promote integration of the LTR retrotransposons Ty1 and Ty3 in Saccharomyces cerevisiae, indicating that the contribution of host factors to integration is common among distantly related organisms. The DNA repair factors are of particular interest because they may identify the pathways that repair the single-stranded gaps opposite integration sites of LTR retroelements.
Retrotransposon Tf1 induces genetic adaptation to environmental stress.
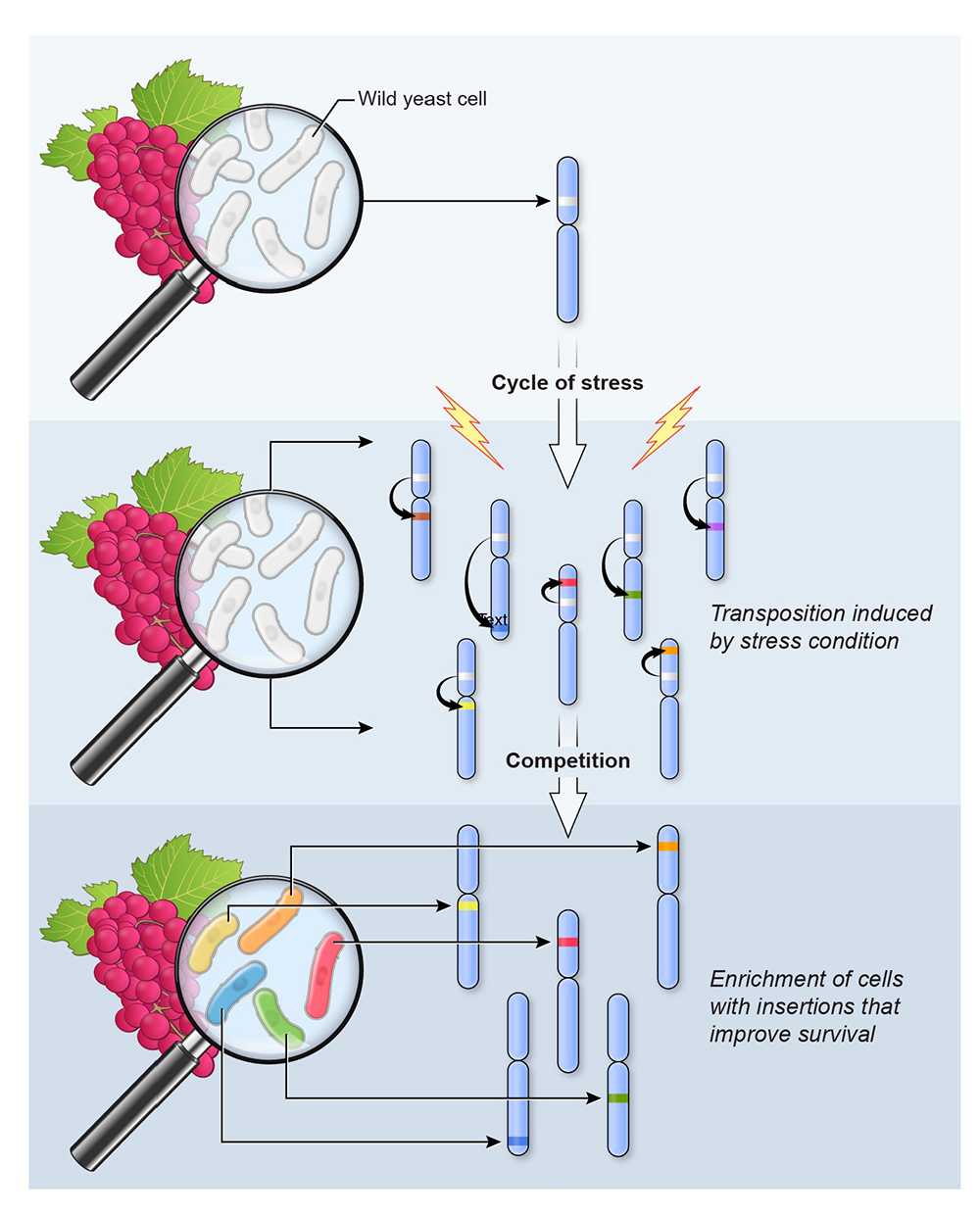
Ever since Barbara McClintock discovered transposable (“Controlling”) elements in maize, it has been accepted that they are activated by changes in environmental conditions. Although increased mobility has long been thought to benefit the host, the precise impact and importance of this activity has not been directly studied. Schizosaccharomyces pombe possesses a compact genome that tightly restricts retrotransposon expression under normal growth conditions. However, when the retrotransposon Tf1 is expressed, it integrates into promoters of RNA Pol II–transcribed genes and, in many cases, this increases transcription of adjacent genes. This finding, together with the Tf1 preference for stress-response promoters, led to the idea that Tf1 could be beneficial to its host by creating a pool of new insertions that improve survival of environmental stress. We tested the hypothesis by studying the fitness of cells with genomic insertions of Tf1 when exposed to stress. Diverse cultures containing Tf1 integrated at 42,000 positions were grown competitively in cobalt. The proportion of cells with Tf1 at 141 positions greatly increased, suggesting that the integrations improved growth in cobalt. Analysis of the positions and reconstruction of strains with single insertions indicate that Tf1 integration improved growth in cobalt by inducing key regulators of the cell-regulatory TOR pathway. A critical feature of Tf1 activity and that of the closely related Tf2 is that their promoters participate in the core stress response, showing significant activation when cells are exposed to these stresses. Significantly, we found that the activation of Tf1 transcription results in increased integration frequencies. As a result, each of these features—increased mobility, targeting of promoters, and the stimulation of adjacent genes—promotes adaptation. Having observed these properties, we propose that Tf1 is a highly evolved mutagenic system that benefits the host by driving adaptation to environmental insults. In a model for adaptation through stress-induced mobilization of transposable elements, we propose that repeated exposure to stress results in cycles of increased transposition. Competition between cells containing new insertions results in the selection of sets of insertions that improve survival (Figure 2). An intriguing additional possibility is that, through continued exposure to an unfamiliar stress, several insertions could accumulate in individual cells that together could form the foundation of a new gene-regulatory network (GRN). Such networks would be specific depending on the nature of the existing insult. The assembly of GRNs resulting from integration activity of TEs is a compelling model for how regulatory sequences of TEs have undergone wide-spread domestication in controlling GRNs. Supporting this model is our study of polymorphic Tf1 and Tf2 LTRs present in 57 wild isolates of S. pombe. The enrichment of LTRs in the promoters of heat-shock and sporulation genes provided evidence that TEs do promote adaptation in natural conditions. Together, our results indicate that integration activity provides substantial benefit when cells are subjected to stress.
Click image to view.
Figure 2. Model for adaptation through stress-induced mobilization of transposable elements
Repeated exposure to stress results in cycles of increased transposition. Competition between cells containing new insertions results in the selection of sets of insertions that improve survival.
LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced gene.
The promise of immunotherapy of cancer using gene therapy relies on retroviral vectors to stably integrate the corrective/therapeutic sequences in the genomes of the patient’s cells. First-generation gene therapy used vectors derived from gamma retroviruses that were successful in correcting X-linked severe combined immunodeficiency (SCID-X1). However, the integration pattern had a bias for promoter sequences that resulted in the activation of proto-oncogenes and progression to T cell leukemia. Such adverse outcomes led to the use of lentivirus vectors for more recent gene-therapy treatments. This switch to HIV-1–based vectors has occurred despite a fundamental lack of information about integration levels at specific genes, including at proto-oncogenes. Structural and biochemical data show that HIV-1 integrase (IN) interacts with the host factor LEDGF/p75 (a chromatin-binding protein and transcription coactivator), and that the interaction favors integration in the actively transcribed portions of genes (transcription units). However, little is known about how LEDGF/p75 recognizes transcribed sequences and whether cancer genes are favored.
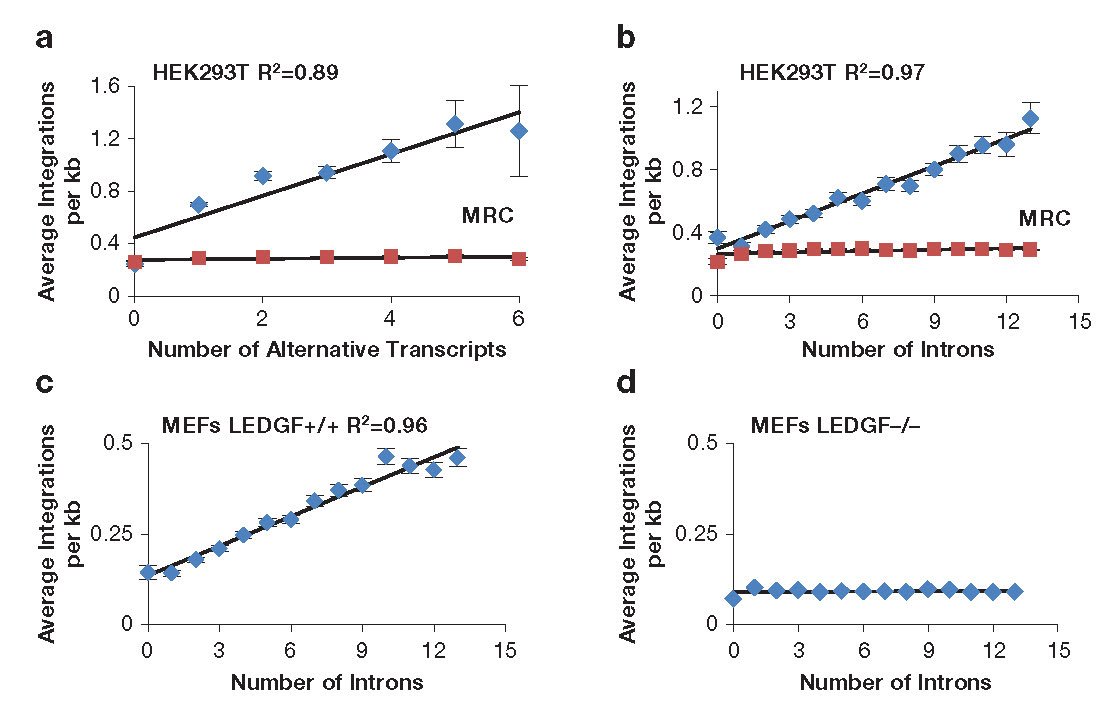
To measure integration levels in individual transcription units and to identify the determinants of integration-site selection, we generated a high-density map of the integration sites of a single-round HIV-1 vector in HEK293T tissue culture cells [Reference 3]. Improvements in sequencing methods allowed us to map 961,274 independent integration sites; most of the sites occurred in just 2,000 transcription units. Importantly, the 1,000 transcription units with the highest numbers of integration sites were highly enriched for cancer-associated genes, which raised concerns about the safety of using lentivirus vectors in gene therapy. Analysis of the integration site densities in transcription units (integration sites per kb) revealed a striking bias that favored transcription units that produced many spliced mRNAs and with transcription units that contain high numbers of introns (Figures 3A and 3B) [Reference 3]. The correlations were independent of transcription levels, size of transcription units, and length of the introns. Analysis of previously published HIV-1 integration site data showed that integration density in transcription units in mouse embryonic fibroblasts also correlated strongly with intron number and that the correlation was absent from cells lacking LEDGF (Figures 3C and 3D). The data suggest that LEDGF/p75 not only tethers HIV-1 integrase to the chromatin of active transcription units but also interacts with mRNA splicing factors. To test this, our collaborators Matthew Plumb and Mamuka Kvaratskhelia used tandem mass-spectrometry (MS-MS) to identify cellular proteins from nuclear extracts of HEK293T cells that interacted with GST-LEDGF/p75 (LEDGF/p75 tagged with glutathione S-transferase). The proteomic experiments found that LEDGF/p75 interacted with many components of the splicing machinery, including the small nuclear ribonucleic proteins (snRNP) SF3B1, SF3B2, and SF3B3 of U2 (a small nuclear RNA component of the spliceosome), U2–associated proteins PRPF8 and U2SURP, a factor of the U5 snRNP (SNRNP200), and many hnRNPs (heterologous ribonucleoproteins) that are associated with alternative splicing. The broad range of interactions with splicing factors suggested that LEDGF/p75 might contribute to splicing reactions. To test this, we performed RNAseq on HEK293T cells that were altered with TALEN endonucleases to truncate or delete PSIP1, the gene encoding LEDGF/p75. Analysis of transcription units that produced two or more spliced mRNA products showed that bi-allelic deletion of LEDGF/p75 significantly changed the ratio of spliced products in large numbers of transcription units. These results, together with our finding that integration in highly spliced transcription units was dependent on LEDGF, provide strong support for a model in which LEDGF/p75 interacts with splicing machinery and directs integration to highly spliced transcription units.
Click image to view.
Figure 3. Integration density in transcription units correlates with amounts of splicing.
The numbers of HIV-1 integrations per kb in transcription units correlates with the amount of splicing (a and b). The preference for highly spliced transcription units depends on host factor LEDGF (c and d). MEFs: mouse embryonic fibroblasts; MRC: Matched Random Control.
Additional Funding
- NIH Intramural AIDS Targeted Antiviral Program (2017 and 2018)
Publications
- Hickey A, Esnault C, Majumdar A, Chatterjee A, Iben J, McQueen P, Yang A, Mizuguchi T, Grewal S, Levin HL. Single nucleotide specific targeting of the Tf1 retrotransposon promoted by the DNA-binding protein Sap1 of Schizosaccharomyces pombe. Genetics 2015;201:905-924.
- Rai S, Sangesland M, Esnault C, Lee M, Chatterjee A, Levin HL. Host factors that promote retrotransposon integration are similar in distantly related eukaryotes. PLoS Genetics 2017;13:p1006775.
- Singh PK, Plumb MR, Ferris AL, Iben JB, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH, Kvaratskhelia M, Levin HL. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev 2015;29:2287-2297.
- Rai SK, Atwood-Moore A, Levin HL. Duplication and transformation of the Schizosaccharomyces pombe collection of deletion strains. Methods Mol Biol 2018;1721:197-215.
- Rai SK, Atwood-Moore A, Levin HL. High-frequency lithium acetate transformation of Schizosaccharomyces pombe. Methods Mol Biol 2018;1721:167-177.
Collaborators
- Shiv Grewel, PhD, Laboratory of Biochemistry and Molecular Biology, NCI, Bethesda, MD
- Stephen Hughes, PhD, Retroviral Replication Laboratory, HIV Drug Resistance Program, NCI, Frederick, MD
- Mamuka Kvaratskhelia, PhD, Ohio State University, Columbus, OH
- Philip McQueen, PhD, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- Matthew Plumb, BS, Ohio State University, Columbus, Ohio
Contact
For more information, email henry_levin@nih.gov or visit http://sete.nichd.nih.gov.


