Assembly and Function of Drosophila Chromatic Circuits

- Chi-Hon Lee, MD, PhD, Head, Section on Neuronal Connectivity
- Chun-Yuan Ting, PhD, Staff Scientist
- Yang Li, PhD, Visiting Fellow
- Jiangnan Luo, PhD, Visiting Fellow
- Bo-mi Song, PhD, Intramural Research Training Award Fellow
- Bo Shi, MSc, Graduate Student
- Moyi Li, BA, Biological Laboratory Technician
Mapping color-vision circuits
Visual animals utilize spectral information in two ways: true color vision, which differentiates spectral compositions largely independent of brightness, allows animals to recognize objects and register and retrieve color memory; innate spectral preference, which depends on intensity but not learning, often reflects individual species’ specific ecological needs. Using a combination of genetic, histological, electrophysiological, imaging, and behavioral approaches, our group studies how visual circuits process chromatic information to guide behaviors in Drosophila. Our strategy is to (1) identify key neuronal types and map their synaptic connections, (2) examine functional requirements of identified neurons for color vision and spectral preference behaviors, and (3) determine the synaptic mechanisms that transform visual signals at different processing stages.
Using molecular genetics, histology, and serial–EM (electron microscope) reconstruction, we mapped the synaptic circuits of the chromatic photoreceptors R7s and R8s and their synaptic targets, the Tm and Dm neurons, in the peripheral visual system, as well as their downstream targets in the higher visual system, the lobula. The medulla projection (Tm) neurons (Tm5a/b/c, Tm9, and Tm20), which are analogous to vertebrate retinal ganglion cells, relay photoreceptors to higher visual centers, while the Dm (Dm8) neurons provide an indirect pathway by relaying photoreceptors to Tm neurons. To probe the synaptic connections between these neurons, we developed several versions of GRASP (GFP reconstitution across synaptic partners) methods to map active synapses. Using such methods, we found that the chromatic photoreceptors R7 (UV–sensing) and R8 (blue/green-sensing) provide inputs to a subset of first-order interneurons. Tm9/20/5c and Tm5a/b receive direct synaptic inputs from the retinotopic R8s and R7s, respectively, consistent with their functions in processing single visual-pixel information. In contrast, the amacrine neuron Dm8 pools inputs from 14 R7s and provides input for Tm5c. Based on histology, we found that the Tm5a/b/c and Tm20 neurons relay photoreceptor signals to the lobula, the higher visual center, which, in insects, has been implicated in processing and relaying color information to the central brain. Using the GRASP method and a series of lobula neuron drivers, we identified the lobula neurons that receive direct synaptic inputs from the chromatic Tm neurons Tm5a/b/c and Tm20. We found four types of lobula neurons that form synaptic contacts with chromatic Tm neurons: two novel lobula intrinsic neurons, Li3 and Li4, and two lobula projection neurons, LT11 and LC14. Each LT11 elaborates a large dendritic tree to cover Lo4–6, the entire lobula layers, and projects an axon to optic glomeruli in the central brain. Each Li4 extends dendrites to cover Lo5, about 60% of the lobula layer, while Li3 dendrites cover about 15% of lobular layers 5 and 6. To confirm synaptic connections observed at the light-microscopic level, we developed a two-tag EM double-labeling technique that highlights both presynaptic and postsynaptic terminals in the same preparation. By combining two orthogonal expression systems and two different peroxidases, HRP (horse radish peroxidase) and APX (ascorbate peroxidase), we expressed HA–tagged mitochondrion-targeted APX in Tm5c and membrane-tethered HRP in their LT11 postsynaptic neurons, thereby confirming appositions among synaptic profiles. Our anatomical study revealed that the lobula neurons integrate multiple chromatic inputs from Tm neurons over a large receptive field, suggesting that color vision is of low spatial resolution.
To identify neurons that are involved in innate spectral preference, we used genetic methods to systematically inactivate different first-order interneurons and then examined their behavioral consequences. We had previously found that the amacrine Dm8 neurons, which receive UV–sensing R7 photoreceptor inputs, are both required and sufficient for animals’ innate spectral preference for UV light. Using behavioral assays, we further found that inactivating Tm5c, one of Dm8’s synaptic targets, abolishes UV preference, establishing Tm5c as the key downstream target for spectral preference. Using single-cell transcript profiling and immunohistochemistry methods, we found that both Dm8 and Tm5c express the vesicular glutamate transporter (VGlut) and that Tm5c expresses four kainite-type glutamate receptors (Clumsy, DKaiR1C, DKaiR1C, and CG11155). RNAi-knockdown of the receptors in Tm5c, which prevents Tm5c from receiving Dm8 inputs, abolished UV preference, indicating that the receptors are functionally required in Tm5c for UV preference behaviors. Thus, the R7s→Dm8→Tm5c connections constitute a hard-wired glutamatergic circuit for detecting dim UV light. To determine the functional properties of these CNS glutamate receptors, we reconstituted DKaiR1D in oocytes and determined its electrophysiological properties. We found that DKaiR1D forms a rapidly activating and desensitizing homotetramer receptor that is glutamate-gated and calcium-permeable. Notably, DKaiR1D is inhibited by NMDA and AP5, the respective agonist and antagonist of vertebrate NMDA receptors, indicating that insect glutamate channels have a very different pharmacology from that of vertebrates. Two-photon calcium imaging of Dm8 and Tm5c expressing the calcium indicator GCaMP6f revealed that the activity of both neurons is suppressed by full-field UV light illumination. The light responses of Dm8 depend on R7 activity and Dm8’s expression of the histamine chloride channel ORT, indicating that Dm8 receives inhibitory signals from photoreceptors via ORT and conveys sign-inverted signals to Tm5c via kainate receptors.
In contrast to UV preference, color vision in Drosophila is mediated by several partially redundant pathways. To identify color-vision circuits, we developed a novel aversive operant conditioning assay to test animals’ ability for intensity-independent color discrimination. We successfully trained single flies to discriminate between equiluminant blue or green stimuli. We found that wild-type flies can be trained to avoid either blue or green, while mutants lacking functional R7 and R8 photoreceptors cannot, indicating that the color entrainment requires the function of the narrow-spectrum photoreceptors R7s and/or R8s. Furthermore, inactivating four types of first-order interneurons, Tm5a/b/c and Tm20, abolishes color learning, but inactivating different subsets of the neurons is insufficient to block color learning. In summary, our results suggest that true color vision is mediated by parallel pathways with redundant functions. The apparent redundancy in learned color discrimination is in sharp contrast to innate spectral preference, which is mediated by the single pathway R7s→Dm8→Tm5c.
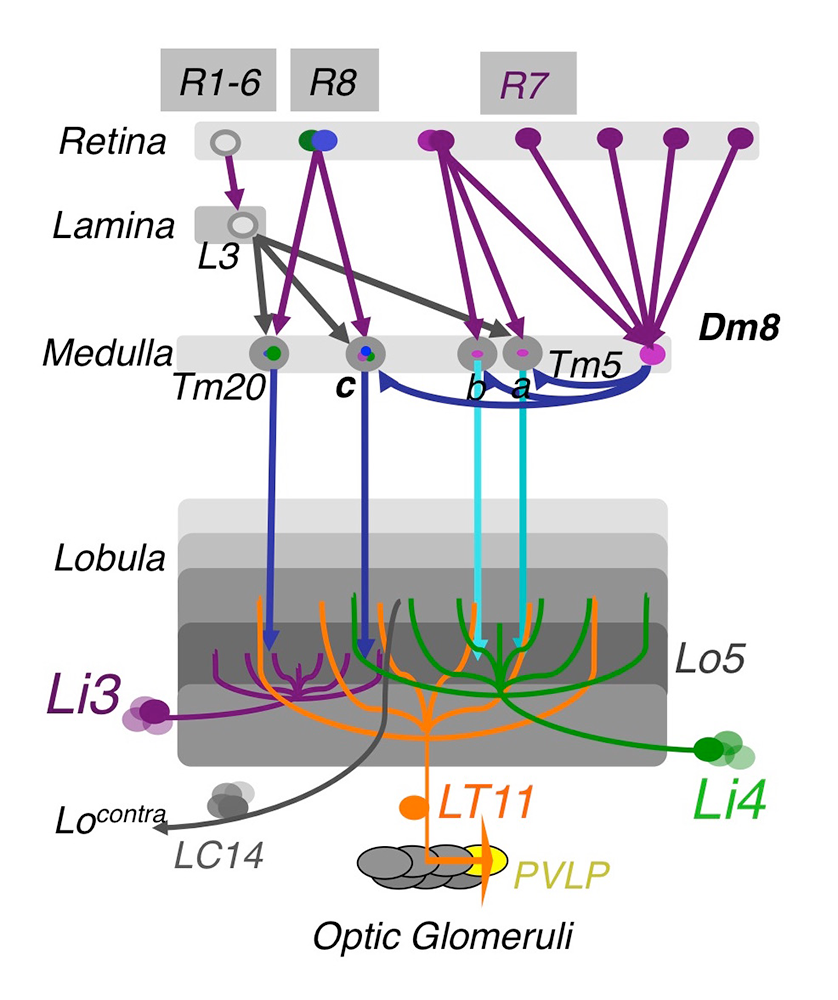
Click image to view.
Figure 1. The chromatic circuit in Drosophila
Fly vision is mediated by three types of photoreceptors, R1–6, R7, and R8, each responding to a specific spectrum of light and connecting to different synaptic partners in the lamina and medulla neuropils. The chromatic photoreceptors R7 and R8 provide inputs for the amacrine neuron Dm8 and the transmedulla neurons Tm5a/b/c and Tm20. The transmedulla neurons transmit visual signals to four types of lobular neurons, LT11, LC14, Li3, and Li4, in the higher visual center.
Dendritic development of Drosophila optic lobe neurons
Correct brain wiring requires axons and dendrites to be routed to appropriate regions, such as layers and columns, in order for correct synaptic connections to be formed during development. Many neuropsychiatric disorders, such as Down syndrome, Fragile X syndrome, and Rett syndrome, have developmental origins and exhibit defects in dendritic morphology, such as changes in branching numbers and patterns. Dendritic defects could cause neuronal connectivity defects, which likely underlie neurological and cognitive deficits. It remains unclear, however, how genetic disorders lead to dendritic patterning defects during development, which in turn lead to erroneous connections and functional deficits in adults.
Our group uses Drosophila optic lobe neurons as a model in which to study dendritic development and neural circuit assembly in the central nervous system. Similar to the vertebrate cortex and retina, the Drosophila optic lobe is organized in columns and layers, suggesting that fly visual neurons and vertebrate cortex neurons face similar challenges in routing their dendrites to specific layers and columns during development. Furthermore, fly visual neurons have unique advantages. First, the medulla neurons extend dendrites to form synapses in a lattice-like structure. Second, specific subtypes of medulla neurons can be specifically labelled and their genetics manipulated at single-cell resolution. Third, the synaptic circuits have been characterized at the ultrastructural level and can be analyzed at the light-microscopic level. Forth, functional deficits can be fully characterized using behavioral assays and functional imaging assays. We further developed several novel techniques to generate high-resolution images, to standardize and compare dendritic patterns, and to visualize synaptic connections at the light- and electron-microscopic levels, thus facilitating phenotypic analyses.
We carried out two genetic screens to identify molecular determinants that control dendritic patterning of Tm20 and Dm8 neurons. We focused on four types of dendritic developmental defects in: (1) the initiation of main dendritic branches; (2) the dendritic projection directions; (3) the layer-specific targeting of dendrites; and (4) dendritic field sizes. From the genetic screens, we identified adhesion receptors, morphogen receptors, signaling molecules, and cytoskeletal regulators that are cell-autonomously required in Tm20 and/or Dm8 neurons for proper dendritic development. The RNAi screen identified families of cadherins (calcium-dependent cell-adhesion proteins) and cadherin-like receptors that are required for proper initiation of dendritic branches and receptive field sizes. N-cadherin, the classical cadherin, is required cell-autonomously in Tm20 neurons for layer-specific initiation of main dendritic branching points. Unlike wild-type Tm20 neurons, which extended most dendritic branches from one or two primary branching nodes located in the medulla M3 layer, Ncad–mutant Tm20 neurons initiated the main dendritic branches in the M2 layers. The layer shift of the main branching nodes in Ncad–mutant Tm20 is further compounded with an alteration in layer-specific targeting of their dendritic arbors and their planar projection directions. Interestingly, the total dendritic length was unaffected, suggesting that the Ncad mutation specifically affects the initiation of primary dendritic branches rather than branch trimming.
The loss-of-function mosaic screen identified two pathways that regulate the sizes of dendritic trees. We previously demonstrated that the TGF-beta/Activin signaling pathway negatively controls the sizes of the dendritic fields of Tm20 and Dm8 neurons. Mutant Tm20 neurons lacking Activin signaling components, such as the receptor Baboon and the downstream transcription factor Smad2, elaborated an expanded dendritic tree spanning several medulla columns. Morphometric analyses revealed that baboon and smad2 mutations significantly reduce dendritic termination frequency but not branching frequency. Using a modified GRASP method that we had developed, we found that the expanded dendritic tree of a mutant Tm20 neuron forms aberrant synaptic contacts with several neighboring R8 photoreceptors. RNAi knock-down of Activin in R7 and R7 further showed that Activin derived from photoreceptors R7 and R8 acts at short ranges on R7’s and R8’s respective synaptic targets, Dm8 and Tm20. Recently, we found that the insulin signaling pathway positively regulates the dendritic tree size of Dm8. Mutant Dm8 neurons lacking insulin receptor or the downstream signaling components TOR or Rheb have a small dendritic tree. On the other hand, mutant Dm8 neurons lacking the negative regulators of the insulin signaling pathway, Pten or TSC1, have an abnormally expanded dendritic tree compared with wild-type neurons. Mis-regulation of TOR signaling, collectively called mTORopathies, has been implicated in several focal malformations of cortical development (MCD) subtypes associated with epilepsy and dendritic morphological defects. However, the dissection of TOR signaling pathways in complex mammalian nervous systems has been difficult. The Dm8 and Tm20 dendritic development system we developed thus provides a swift way to dissect the complex phenotypes of the TOR pathway and to determine the cell-autonomous functions of TOR signaling at the single cell resolution.
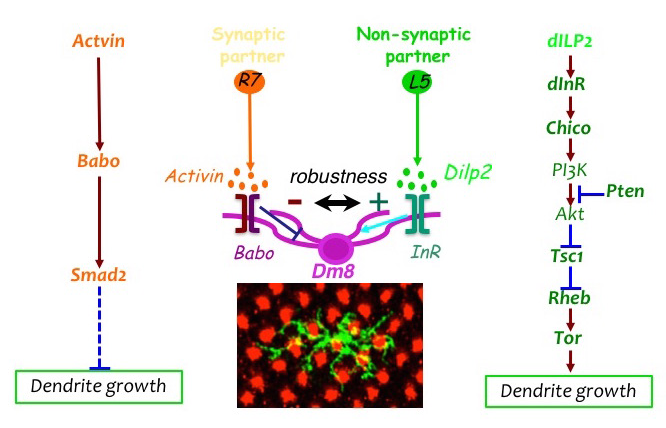
Click image to view.
Figure 2. The afferent-derived factors, Activin and insulin-like peptide, antagonistically regulate the size of dendritic field of the amacrine neuron Dm8.
Two afferent-derived factors, Activin and Drosophila insulin-like peptide2 (Dilp2), signal on the first-order interneuron Dm8 to control the sizes of its dendritic field. Activin is provided by R7 photoreceptors, the synaptic partners of Dm8, while Dilp2 is provided by lamina neurons (LNs), non-synaptic partners of Dm8. Babo: Baboon (Activin receptor); InR: insulin receptor.
Publications
- Biswas TM, Lee C-H. Visual motion: cellular implementation of a hybrid motion detector. Curr Biol 2017;27:R274-276.
- Li Y, Dharkar P, Han T-H, Serpe M, Lee C-H, Mayer ML. Novel functional properties of Drosophila CNS glutamate receptors. Neuron 2016;92:1036-1048.
- Lin T-Y, Luo J, Shinomiya K, Ting C-Y, Lu Z, Meinertzhagen IA, Lee C-H. Mapping chromatic pathways in the Drosophila visual system. J Comp Neurol 2016;524:213-227.
- Luo J, McQueen PG, Shi B, Lee C-H, Ting C-Y. Wiring dendrites in layers and columns. J Neurogenet 2016;30:69-79.
- Kulkarni A, Ertekin D, Lee C-H, Hummel T. Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. eLife 2016;5:e13715.
Collaborators
- Mary Lilly, PhD, Section on Gamete Development, NICHD, Bethesda, MD
- Mark Mayer, PhD, Laboratory of Cellular and Molecular Neurophysiology, NICHD, Bethesda, MD
- Matthew McAuliffe, PhD, Division of Biomedical Imaging Research Services Section, CIT, NIH, Bethesda, MD
- Philip McQueen, PhD, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- Ian Meinertzhagen, PhD, DSc, Dalhousie University, Halifax, Canada
- Kate O'Connor-Giles, PhD, University of Wisconsin, Madison, WI
- Nishith Pandya, BA, Division of Biomedical Imaging Research Services Section, CIT, NIH, Bethesda, MD
- Thomas Pohida, MSEE, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Randy Pursley, MSEE, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Mihaela Serpe, PhD, Section on Cellular Communication, NICHD, Bethesda, MD
- Mark Stopfer, PhD, Section on Sensory Coding and Neural Ensembles, NICHD, Bethesda, MD
- Benjamin White, PhD, Laboratory of Molecular Biology, NIMH, Bethesda, MD


