Deciphering the Virulence Mechanisms of Microbial Pathogens

- Matthias Machner, PhD, Head, Section on Microbial Pathogenesis
- Devanand Bodage, PhD, Visiting Fellow
- Katherine Bonnington, PhD, Visiting Fellow
- Nicole Ellis, PhD, Visiting Fellow
- Pei-Chung Lee, PhD, Visiting Fellow
- Stephanie Lehman, PhD, Visiting Fellow
- Xiao Li, PhD, Visiting Fellow
- Kathryn Johnston, BS, Postbaccalaureate Student
- Mitchell Lee, BS, Postbaccalaureate Student
Our main research goal is to obtain mechanistic insight into the virulence strategies of microbial pathogens. As a model organism, we use the bacterium Legionella pneumophila, the causative agent of a potentially fatal respiratory infection known as Legionnaires' disease. According to the CDC (Centers for Disease Control), the number of Legionnaires' disease cases in the U.S. has risen more than four-fold over the past 15 years, making L. pneumophila an emerging pathogen of increasing relevance. Contrary to what its name may imply, Legionnaires’ disease occurs in individuals of all ages, including children who receive respiratory therapy, newborns who had recently undergone surgery or under-water birth, and children who are immune-compromised. We are committed to an in-depth analysis of the mechanisms that allow L. pneumophila to exploit the human host and cause disease. Insights gained from these studies will ultimately improve our ability to diagnose, prevent, and fight Legionnaires’ disease and related illnesses, thereby contributing to the success of the NICHD’s mission.
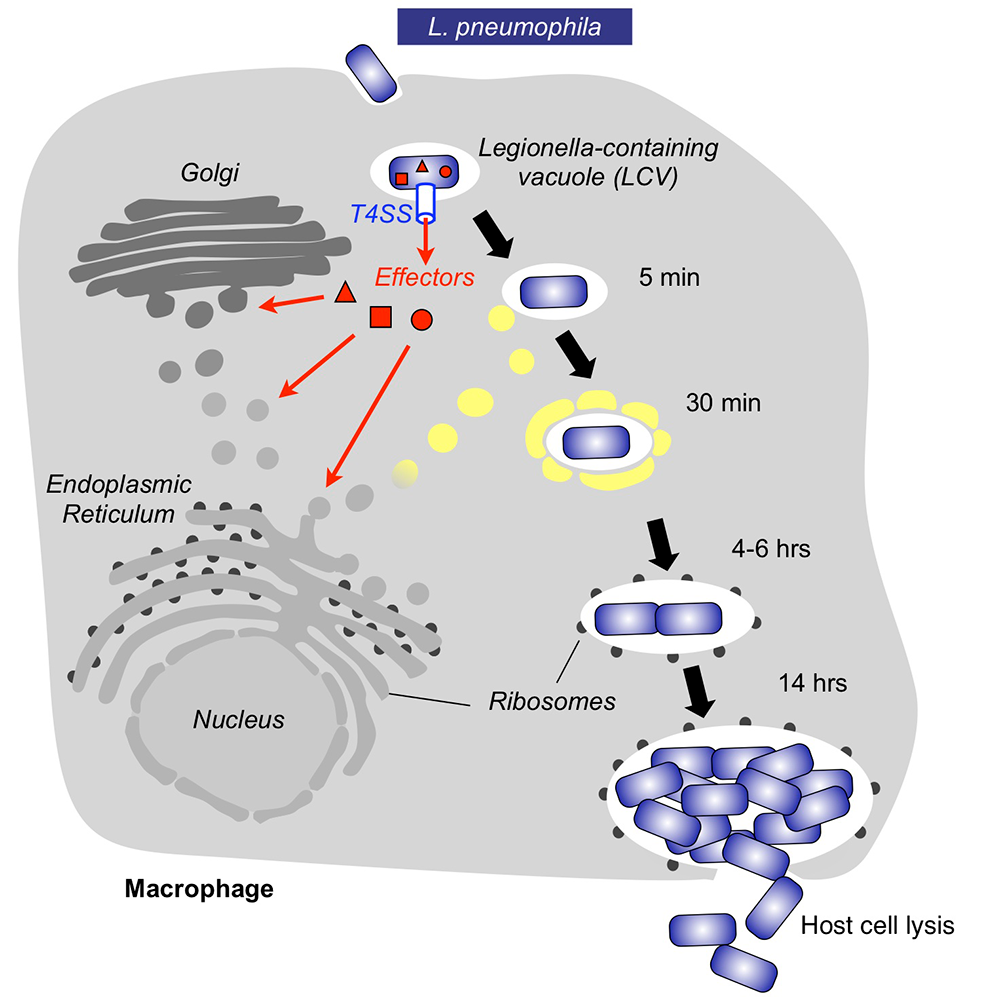
Upon inhalation of contaminated water droplets, L. pneumophila enters the lung and is phagocytosed (taken up) by specialized immune cells known as alveolar macrophages (Figure 1). Instead of being degraded by these cells, the pathogen establishes a protective membrane compartment: the Legionella-containing vacuole (LCV). Within this intravacuolar niche, L. pneumophila can replicate to high numbers before killing the host cell and infecting neighboring cells. The virulence of L. pneumophila relies on the activity of close to 300 proteins, or effectors, that are delivered into the host cytosol by a specialized translocation apparatus called the Dot/Icm type IV secretion system (T4SS) (Figure 1). L. pneumophila mutants with a non-functional T4SS are degraded by macrophages, underscoring the importance of the translocated effectors for host-cell manipulation and bacterial virulence.
Click image to view.
Figure 1. Intracellular replication cycle of Legionella pneumophila
Upon uptake by a macrophage, L. pneumophila delivers a large number of effector proteins (red) through the Dot/Icm type IV secretion system (T4SS) into the host cytosol. The effectors manipulate signaling and trafficking pathways in order to establish conditions favorable for L. pneumophila growth. Eventually, the host cell is lyzed, and L. pneumophila bacteria infect neighboring cells.
Our main research objective is to obtain detailed mechanistic insight into the regulation and function of L. pneumophila effectors by investigating host-pathogen interactions at a molecular, cellular, and structural level. Deciphering the virulence program of this emerging pathogen will set the stage for the development of novel therapeutics aimed at treating or preventing Legionnaires' disease and related illnesses.
A bacterial E3 ligase relic hijacks host-cell ubiquitination.
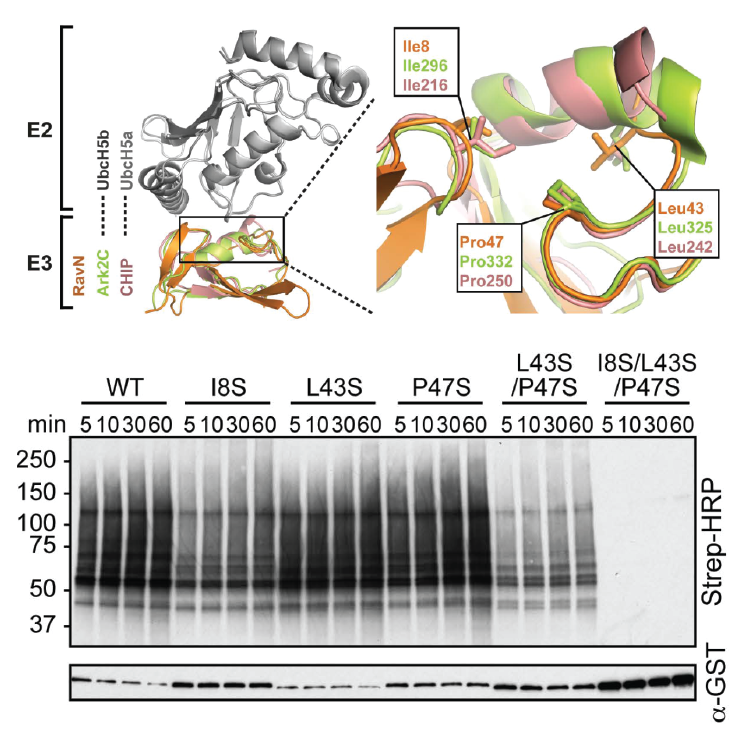
Bacterial pathogens often target conserved host pathways by encoding proteins that are molecular mimics of cellular enzymes, thus tricking the host cell into surrendering its resources to the bacterium. We discovered that L. pneumophila uses such a strategy to exploit ubiquitination, a conserved post-translational modification that is mediated by a family of enzymes called E3 ubiquitin ligases. L. pneumophila encodes its own molecular mimics of E3 ligases, including the effector protein RavN, thereby subverting the ubiquitin pathway for its own benefit during infection. By testing truncated RavN variants in an in vitro reconstitution assay, we found that the E3 ligase activity of RavN is located within its N-terminal region. Using protein crystallography, we found that the crystal structure of RavN has only residual resemblance to conventional eukaryotic U-box ubiquitin ligases (Figure 2), yet its mode of interaction with E2 enzymes has been preserved throughout evolution, indicating that RavN is a ubiquitin ligase relic. In support of this conclusion, we demonstrated that substitution of residues within the predicted E2 binding interface rendered RavN inactive. The study confirmed that, despite having undergone extensive evolutionary changes, L. pneumophila effectors have retained features that are critical for their biological function, including the ability to hijack host factors that are part of the ubiquitylation machinery.
Click image to view.
Figure 2. RavN is a ubiquitin ligase relic from L. pneumophila.
Upper panel. Superposition of RavN and two related E3s in complex with their respective E2s (gray). A close-up view highlights binding-site residues.
Lower panel. Strep–HRP immunoblot to detect poly-ubiquitylation (indicated by the dark smearing pattern) catalyzed by purified RavN or RavN mutants in which binding site residues have been substituted with serine.
Discovery of inhibitors of the Legionella type IV secretion system
Most classical antibiotics kill bacteria or inhibit their growth by disrupting key aspects of their physiology. Coupled with over-prescription of antibiotics and patient non-compliance, this has led to the rapid emergence of multidrug-resistant strains that have become insensitive to the microbicidal or microbiostatic activity of existing compounds. In addition, recent insight into the complexity and sensitivity of the human microbiome and its importance for human health has raised concerns about the excessive use of antibiotics and their collateral effect on commensal microflora. Thus, there is an urgent need for the development of 'smarter' therapeutics that discriminate between pathogens and commensals by selectively targeting virulence components of microorganisms.
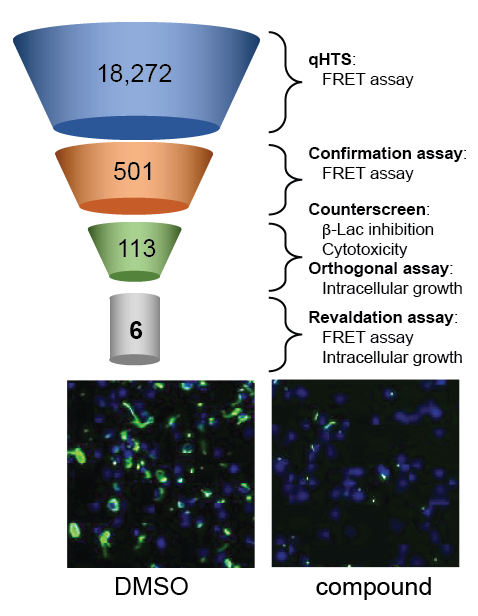
Given their essential role in virulence, bacterial secretion systems represent a compelling target for the development of novel therapeutic agents. In collaboration with Anton Simeonov, we designed a high-throughput fluorescence resonance energy transfer (FRET)–based beta-lactamase (B-Lac) reporter assay and screened a library of over 18,000 compounds for candidates that interfere with the ability of L. pneumophila to deliver a B-Lac-reporter protein into mouse macrophages (Figure 3). Upon vetting 501 candidate compounds in a variety of in vitro and cell-based secondary screens, we identified six lead compounds that fulfilled all criteria of genuine T4SS inhibitors. The compounds efficiently interfered with biological processes that depend on a functional T4SS, such as intracellular bacterial proliferation, but had no detectable effect on L. pneumophila growth in culture medium, conditions under which a T4SS is dispensable. Together, the results suggest that, by directly targeting functional aspects of the T4SS, the six lead compounds render L. pneumophila incapable of using this translocation system to deliver cargo into recipient cells.
Click image to view.
Figure 3. Screen for inhibitors of the L. pneumophila T4SS
Overview of the high throughput screen for compounds that interfere with reporter protein translocation by the T4SS and that, upon addition to macrophage monolayers (blue nuclei), block growth of Lp (green).
A novel platform for the identification of kinase substrates
Eukaryotic cells receive a continuous stream of signals from the intracellular and extracellular environment that are converting into cascades of phosphorylation events that are catalyzed by protein kinases. Kinases covalently transfer the gamma phosphate group of adenosine triphosphate (ATP) onto side chains of substrate proteins, preferentially serine, threonine, or tyrosine residues, thereby altering the activity, localization, or stability of their substrates. Almost all cellular processes are controlled, at least in part, by protein phosphorylation, explaining why mammalian cells encode hundreds of kinases that target thousands of kinase substrates. Not surprisingly, pathogens like L. pneumophila encode molecular mimics of host protein kinases in order to manipulate signaling pathways during infection.
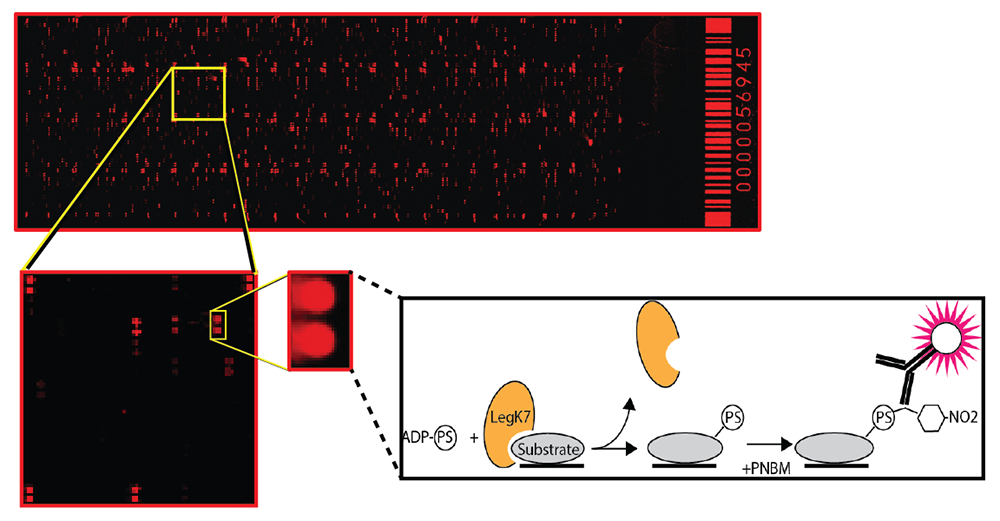
Discovering substrates of bacterial or eukaryotic protein kinases has remained a major challenge because of the transient nature of kinase-substrate interactions and the complexity of the phosphoproteome, even at steady state. To bypass these obstacles, we developed a novel screening platform that combined a previously described thiophosphate labeling technique [Allen JJ et al., Nat Methods 2007;4:511] with a high-density human protein microarray containing more than 9,000 human proteins (Figure 4). As phosphate donor for the kinase reaction, we used adenosine 50-O-(3-thiotriphosphate) (ATPgS). When conjugated onto amino acid residues of substrate proteins on the array, the thiophosphate moiety can be alkylated with p-nitrobenzyl mesylate (PNBM) and detected by a thiophosphate ester–specific antibody followed by a fluorescently labeled secondary antibody. Protein spots on the array that are labeled upon incubation with the wild-type form of a kinase but not its catalytically inactive variant represent putative kinase substrate proteins.
Click image to view.
Figure 4. A novel platform for the identification of kinase substrates
A human protein microarray containing more than 9,000 human proteins (spotted in duplicate) is probed with a kinase (orange color). Substrates proteins (grey color) of the kinase that are phosphorylated on the microarrays are identified using a fluorescently labeled antibody.
The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection.
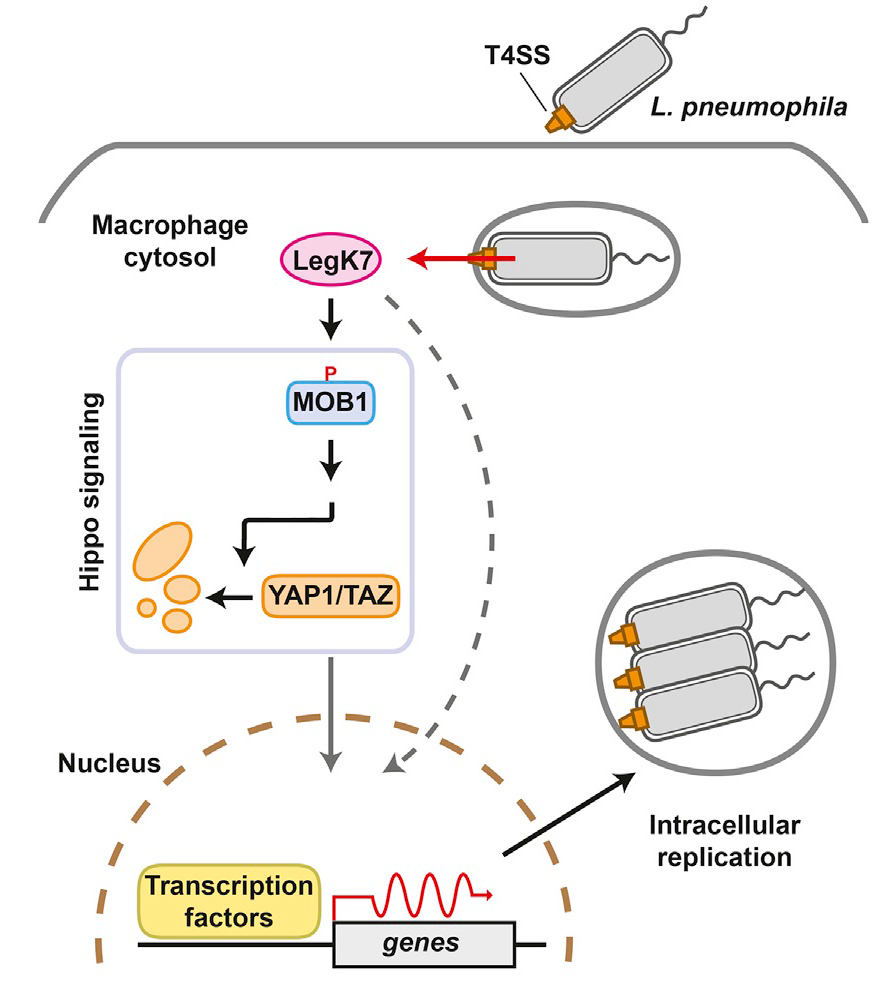
Using Profile Hidden Markov Model–based protein structure prediction to identify cryptic catalytic domains within L. pneumophila effectors, we discovered that the protein LegK7 contains a central domain (residues 183–462) that has folding homology to eukaryotic protein kinases. Using the above-mentioned ATPgS labeling technique, we experimentally confirmed that LegK7 indeed exhibits kinase activity in vitro. Upon probing the protein microarray platform (Figure 4) with LegK7, we identified MOB1 as a direct substrate of LegK7 (Figure 5). MOB1 is a key scaffold protein within the Hippo kinase signaling pathway, which controls cell-cycle progression, cell proliferation, differentiation, and apoptosis in eukaryotes. In a variety of in vitro and cell-based assays, we subsequently showed that LegK7 hijacks the Hippo pathway by molecular mimicking of the host Hippo kinase (MST1 in mammals), which is the key regulator of pathway activation. LegK7, like Hippo/MST1, phosphorylates the scaffolding protein MOB1 on two residues, which triggers a signaling cascade resulting in the degradation of the transcriptional regulators TAZ and YAP1. Transcriptome analysis by RNAseq revealed that LegK7–mediated targeting of TAZ and YAP1 alters the transcriptional profile of mammalian macrophages, a key cellular target of L. pneumophila infection. Specifically, genes targeted by the transcription factor PPARg, which is regulated by TAZ, displayed altered expression, and continuous interference with PPARg activity rendered macrophages less permissive to L. pneumophila intracellular growth (Figure 5). Thus, a conserved L. pneumophila effector kinase exploits the Hippo pathway to promote bacterial growth and infection.
Click image to view.
Figure 5. An important role of LegK7 during L. pneumophila infection
The conserved Hippo pathway controls cell proliferation and development in eukaryotes. LegK7 is an effector kinase in the pathogen L. pneumophila that functionally mimics host Hippo kinase by phosphorylating MOB1. Activated MOB1 triggers degradation of downstream transcriptional regulators, thus altering host gene expression to support bacterial growth.
Publications
- Lee PC, Machner MP. The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection. Cell Host Microbe 2018;24:429-438.
- Romano-Moreno M, Rojas A, Williamson C, Gershlick D, Lucas M, Isupov M, Bonifacino J, Machner MP, Hierro A. Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL. Proc Natl Acad Sci USA 2017;114:E11151-E11160.
- Lin YH, Lucas M, Evans TR, Abascal-Palacios G, Doms AG, Beauchene NA, Rojas AL, Hierro A, Machner MP. RavN is a member of a previously unrecognized group of Legionella pneumophila E3 Ub ligases. PLoS Pathog 2018;14:e1006897.
- Lin YH, Machner MP. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci 2017;130:1985-1996.
Collaborators
- Aitor Hierro, PhD, CIC bioGUNE Institute, Bilbao, Spain
- Michal Jarnik, PhD, Section on Intracellular Protein Trafficking, NICHD, Bethesda, MD
- Joshua LaBaer, MD, PhD, Virginia G. Piper Center for Personalized Diagnostics, Arizona State University, Tempe, AZ
- Anton Simeonov, PhD, Scientific Director, NCATS, Bethesda, MD
Contact
For more information, email machnerm@mail.nih.gov or visit http://machnerlab.nichd.nih.gov.


