Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Neelam Sen, PhD, Research Fellow
- Swati Gaikwad, PhD, Postdoctoral Fellow
- Abdollah Ghobakhlou, PhD, Postdoctoral Fellow
- Suna Gulay, PhD, Postdoctoral Fellow
- Neha Gupta, PhD, Postdoctoral Fellow
- Anil Thakur, PhD, Postdoctoral Fellow
- Vishalini Valabhoju, PhD, Postdoctoral Fellow
- Anil Vijjamarri, PhD, Postdoctoral Fellow
- Fnu Yashpal, PhD, Postdoctoral Fellow
- Quira Zeiden, PhD, Postdoctoral Fellow
- Qiaoyun Zheng, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Laura Marler, BS, Graduate Student
We study the fundamental mechanisms involved in the assembly and function of translation initiation complexes for protein synthesis, using yeast as a model system in order to exploit its powerful combination of genetics and biochemistry. The translation initiation pathway produces an 80S ribosome bound to mRNA, with methionyl initiator tRNA (tRNAi) base-paired to the AUG start codon. The tRNAi is recruited to the small (40S) subunit in a ternary complex (TC) with GTP–bound eukaryotic intitiation factor eIF2 to produce the 43S preinitiation complex (PIC) in a reaction stimulated by eIFs 1, 1A, 3, and 5. The 43S PIC attaches to the 5′ end of mRNA, facilitated by the cap-binding complex eIF4F (comprising eIF4E, eIF4G, and the RNA helicase eIF4A) and poly(A)–binding protein (PABP) bound to the poly(A) tail, and scans the 5′ untranslated region (UTR) for the AUG start codon. Scanning is promoted by eIFs 1 and 1A, which induce an open conformation of the 40S and rapid TC binding in a conformation suitable for scanning successive triplets entering the ribosomal P site (P-out), and by eIF4F and other RNA helicases, such as Ded1, that remove secondary structure in the 5′ UTR. AUG recognition evokes tighter binding of the TC in the P-in state and irreversible GTP hydrolysis by eIF2, dependent on the GTPase–activating protein (GAP) eIF5, releasing eIF2-GDP from the PIC, with tRNAi remaining in the P site. Joining of the 60S subunit produces the 80S initiation complex ready for protein synthesis. Our current aims in this research area are to (1) elucidate functions of eIF1, eIF1A, eIF2, and 40S proteins in TC recruitment and start codon recognition; (2) identify distinct functions of RNA helicases eIF4A (and its cofactors eIF4G/eIF4B), Ded1, and Dbp1, and poly(A)–binding protein (PABP) in mRNA activation, 48S PIC assembly, and scanning in vivo; (3) uncover the mechanisms of translational repression by the repressors Scd6, Pat1, and Dhh1; (4) elucidate possible functions of yeast orthologs of eIF2A and eIF2D in eIF2–independent initiation of translation in stress conditions; (5) elucidate the in vivo functions of Rli1/ABCE1 (translation initiation factor/ATP-binding cassette E1, a ribonuclease inhibitor) and of yeast orthologs of eIF2D and the MCT-1/DENR complex (a translational enhancer) in ribosome recycling at stop codons in vivo.
We also analyze the regulation of amino acid–biosynthetic genes in budding yeast as a means of dissecting fundamental mechanisms of transcriptional control of gene expression. During amino acid limitation, transcription of these genes is coordinately induced by the activator Gcn4 as the result of induction of Gcn4 at the translational level. The eviction of nucleosomes that occlude promoter DNA sequences and block access by RNA polymerase is thought to be a rate-limiting step for transcriptional activation. Previous studies implicated certain histone chaperones, ATP–dependent chromatin-remodeling complexes, or histone acetyltransferase (HAT) complexes in eviction of promoter nucleosomes at certain yeast genes, but it is unclear whether these co-factors function at Gcn4 target genes. Our aim is to elucidate the full set of co-factors that participate in promoter nucleosome eviction at Gcn4 target genes, their involvement in this process genome-wide, and the transcriptional consequences of defective nucleosome eviction. Functional cooperation among the chromatin-remodeling complexes SWI/SNF, RSC, and Ino80, as well as the HAT complexes SAGA, NuA4, NuA3, and Rtt109/Asf1, in these processes are under study. We also recently discovered that Gcn4 can activate transcription from binding sites within the coding sequences (CDS) of its target genes, inducing internal subgenic sense and antisense (AS) transcripts in addition to the conventional full-length transcripts that initiate 5′ of the CDS; and we are probing both the mechanism and possible regulatory functions of these internal AS transcripts.
eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast.
Our previous cryo–EM analysis of partial yeast PICs revealed distinct conformations relevant to different stages of initiation. A py48S–open complex exhibits upward movement of the 40S head from the body that widens both the mRNA binding cleft and the P site and eliminates interactions of the 40S subunit with Met–tRNAi and mRNA that are evident in the py48S–closed complex. The py48S–open conformation seems well suited for scanning of successive triplets for complementarity to Met–tRNAi, with TC anchored in the unstable P-out conformation; whereas py48S–closed exhibits the more stable P-in conformation required for start-codon selection. During the transition from py48S–open to py48S–closed, the unstructured N-terminal tail (NTT) of factor eIF1A assumes a defined structure and deploys five basic residues to interact extensively with the tRNAi anticodon or mRNA nucleotides surrounding the AUG codon or rRNA, thus suggesting that the eIF1A NTT directly stabilizes the P-in state. Interestingly, EIF1AX mutations altering the human eIF1A NTT are recurring mutations associated with uveal melanoma (UM). We found that substituting all five basic residues, and seven UM–associated substitutions, in yeast eIF1A suppresses initiation at near-cognate UUG codons and AUGs in poor sequence context. Ribosome profiling of the UM–associated NTT substitution R13P reveals heightened discrimination against unfavorable AUG context throughout the translatome. Both the R13P and K16D substitutions were shown to destabilize the closed complex at UUG codons in reconstituted PICs. We thus conclude that electrostatic interactions between eIF1A NTT basic residues and nucleotides in tRNAi, mRNA, or rRNA in the decoding center stabilize the closed conformation of the PIC and promote utilization of suboptimal start codons in vivo. We predict that UM–associated EIF1AX mutations alter the expression of human oncogenes or tumor suppressor genes by increasing discrimination against poor initiation codons.
eIF1 Loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex.
As described above, AUG recognition evokes rearrangement from an open PIC conformation with the TC in a P-out state to a closed conformation, with the TC more tightly bound in the P-in conformation. Factor eIF1 binds to the 40S subunit and exerts a dual role of enhancing TC binding to the open PIC conformation while antagonizing the P-in state, necessitating eIF1 dissociation for start codon selection to proceed. Our previous cryo–EM structures of partial yeast PICs revealed juxtaposition of eIF1 Loop 2 with the Met–tRNAi D loop in the P-in state and predict a distortion of Loop 2 from its conformation in the open complex to avoid a clash with Met–tRNAi. We showed that Ala substitutions in Loop 2 increase initiation at both near-cognate UUG codons and AUG codons in poor context in vivo. Consistently, the D71A–M74A double substitution stabilizes TC binding to 48S PICs reconstituted with mRNA harboring a UUG start codon, without affecting eIF1 affinity for 40S subunits. Similar but relatively stronger decreases in discrimination against poor start codons were conferred by arginine substitutions in Loop 2; and none of the Loop 2 substitutions perturbed the rate of TC loading on scanning 40S subunits in vivo. The findings indicate that electrostatic and steric clashing between the eIF1 Loop 2 and tRNAi D loop impede Met–tRNAi accommodation specifically in the P-in state of the closed complex without influencing the P-out mode of TC binding to the open complex; and Arg substitutions convert the Loop 2–tRNAi clash to an electrostatic attraction that stabilizes P-in and enhances selection of poor start codons in vivo. Thus, in contrast to the eIF1A NTT that specifically stabilizes the closed/P-in state of the PIC and enables recognition of poor start codons, eIF1 Loop 2 destabilizes the P-in state and helps ensure relatively greater initiation frequencies for optimal start codons in vivo.
Tma64 (eIF2D), Tma20 (MCT-1), and Tma22 (DENR) recycle post-termination 40S subunits in vivo.
The recycling of ribosomal subunits after translation termination at stop codons is critical for efficient gene expression, as it liberates free ribosomal subunits, deacylated tRNA, and mRNA for use in further rounds of translation. Rli1/ABCE1 catalyzes the first stage of recycling, splitting the 80S ribosome into a free 60S subunit and a tRNA/mRNA–bound 40S subunit. The next step of recycling, dissociation of tRNA and mRNA from the 40S, has been reconstituted in vitro with the single protein ligatin/eIF2D, or the two interacting proteins MCT-1 (also known as MCTS1) and DENR that are homologous to the N- and C-termini, respectively, of eIF2D. Earlier work suggested that the canonical initiation factors eIF1, eIF1A, eIF3, and eIF3j can also recycle 40S post-termination complexes, but it was unknown which, if either, of these redundant recycling mechanisms occurs in living cells. eIF2D was also shown to substitute for eIF2 in tRNAi recruitment for certain mRNAs in vitro, but evidence that eIF2D functions during initiation in vivo was lacking. To address these questions, we performed ribosome profiling on yeast mutants lacking eIF2D (Tma64) together with MCT-1 (Tma20) or DENR (Tma22). Both double mutants revealed that 80S ribosomes queued immediately upstream of stop codons, consistent with a genome-wide block in 40S recycling at stop codons. We also found decisive evidence that the unrecycled 40S complexes in the mutants could reinitiate translation at AUG codons located downstream in the 3′ UTR, as indicated by 80S peaks at such AUG codons in the profiling data, and by detecting expression of epitope-tagged 3′ UTR translation products that was diminished by eliminating the presumptive AUG start codons. In vitro translation experiments using reporter mRNAs containing upstream ORFs (uORFs) further established that reinitiation at coding sequences downstream of the uORFs increased in cell extracts devoid of these proteins. In some cases, 40S ribosomes appeared to rejoin with 60S subunits and undergo an alternative 80S reinitiation process in 3′ UTRs, previously observed in cells depleted of Rli1, that involves unrecycled 80S post-termination complexes. The results support a crucial role for eIF2D (Tma64), MCT-1 (Tma20), and DENR (Tma22) in the recycling of 40S ribosomal subunits at stop codons and thereby diminishing translation reinitiation following termination at both uORFs in 5′ UTRs and at the stop codons of the main coding sequences of most yeast mRNAs.
SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast.
A key unsolved aspect of transcriptional activation by Gcn4 (as well as by other transcription factors) is how it mediates the eviction of the “–1” and “+1” nucleosomes that occlude promoter DNA and block access by general transcription factors (GTFs) and Pol II. We are addressing the mechanism of nucleosome eviction and the consequences of defects in this process on transcription for the hundreds of co-regulated genes in the Gcn4 transcriptome as well as for all other constitutively expressed genes. Previously, we showed, by H3 chromatin immunoprecipitation (ChIP) coupled to deep-sequencing (H3 ChIP-Seq) of wild-type and mutant yeast strains lacking Snf2, Gcn5, and Ydj1, that these cofactors collaborate in evicting H3 from the –1 and +1 nucleosomes and intervening nucleosome-depleted region (NDR) at a large fraction of Gcn4 target genes. Moreover, we found that the cofactors cooperate similarly at the majority of all other promoters. Having found that nucleosome eviction in the induced Gcn4 transcriptome is only partially impaired in cells lacking Snf2, we surmised that SWI/SNF cooperates with one or more other remodeling factors in evicting promoter nucleosomes. Considering that RSC and SWI/SNF belong to the same family of remodeling complexes, we studied whether SWI/SNF and RSC cooperate in nucleosome eviction at genes induced by Gcn4, and also at genes expressed constitutively at high levels where we had previously found that SWI/SNF cooperates with Gcn5 and Ydj1. We also explored whether SWI/SNF resembles RSC in determining the positions of –1 and +1 nucleosomes and hence, NDR width, at highly expressed genes.
Our findings reveal a previously undetected widening of NDRs, in addition to eviction of the –1 and +1 nucleosomes, on induction of Gcn4 target genes in wild-type cells, and demonstrate that SWI/SNF and RSC have distinct and equally critical roles in achieving wide, nucleosome-depleted NDRs for robust transcription at these induced genes. We also uncovered cooperation between SWI/SNF and RSC in nucleosome positioning and eviction at the most highly transcribed subset of constitutively expressed genes, suggesting their general cooperation in achieving high transcription rates. The occupancies by both remodelers were found to be greatest at highly expressed or induced genes, supporting direct functions for both remodelers at this group of genes. Our results reveal an extensive division of labor between SWI/SNF and RSC in promoter nucleosome eviction and displacement at the most highly transcribed genes in yeast.
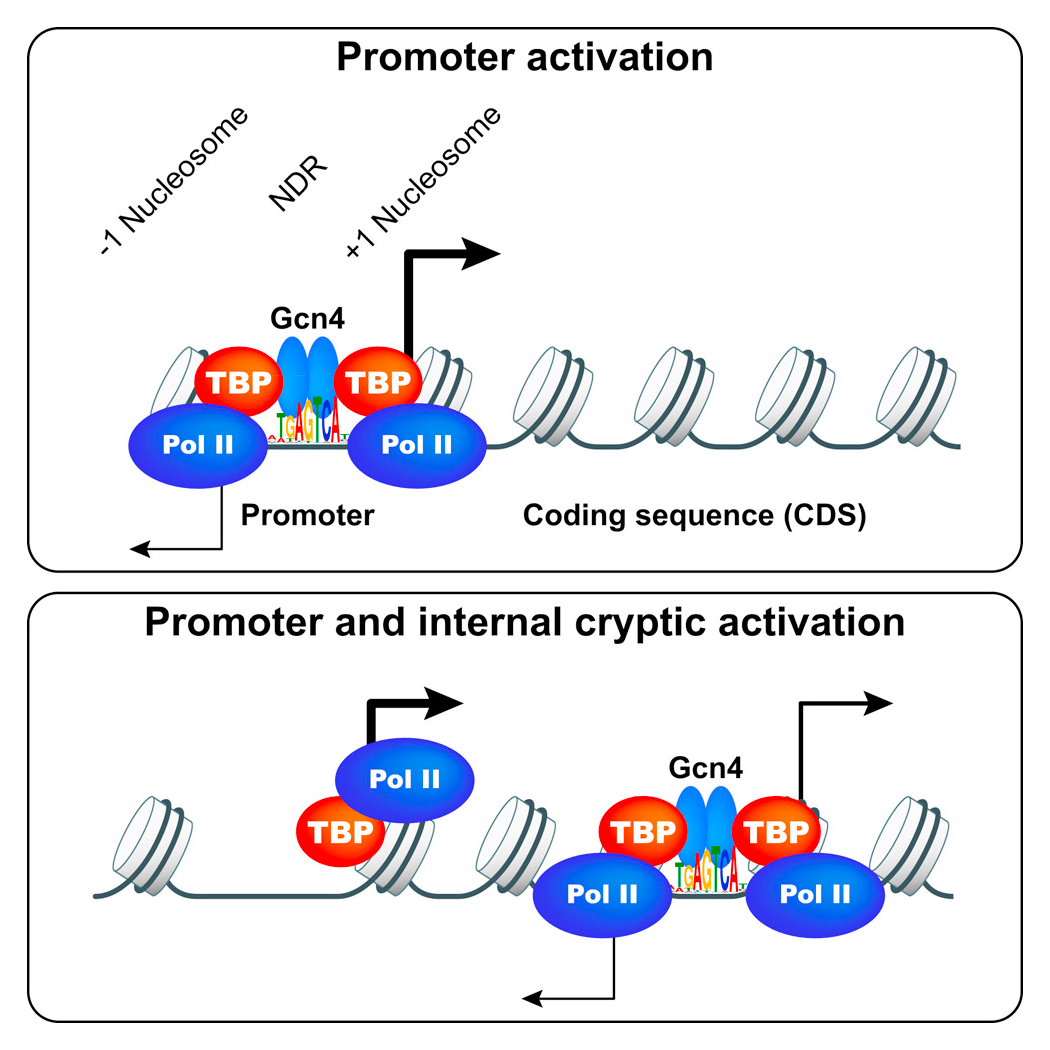
Gcn4 binding within coding regions can activate both internal and canonical 5′ promoters in yeast.
We are also interested in determining the role of promoter nucleosome eviction in controlling binding of Gcn4 itself upstream from the promoters of its target genes, and set out first to define all the binding sites for Gcn4 throughout the genome in wild-type cells. ChIP-seq analysis of Gcn4 binding revealed 546 genomic sites occupied by Gcn4 in starved cells, representing only about 30% of all genomic sequences with significant matches to the consensus Gcn4 binding motif. Analysis of nucleosome occupancy data from MNase-seq analysis revealed that distance of a motif from the nearest nucleosome dyad and its match to the consensus sequence are the major determinants of Gcn4 binding in vivo. Surprisingly, only about 40% of the bound sites are in promoters, and analysis of genome-wide mRNA expression data and ChIP-seq analysis of RNA Pol II in starvation conditions indicates that only about 60% of such promoter-located Gcn4 occupancy peaks activate transcription, revealing extensive negative control over Gcn4 function. Remarkably, most of the remaining around 300 Gcn4–bound motifs reside within coding sequences (CDS), with about 75 representing the only bound motif in the vicinity of a known Gcn4–induced gene. RNA-seq analysis revealed that many such unconventional Gcn4 occupancy peaks map between divergent antisense and sub-genic sense transcripts induced from within CDS under starvation conditions, and are also located adjacent to starvation-induced TBP (TATA-box binding protein) occupancy peaks detected by ChIP-seq analysis. The findings are consistent with Gcn4 activation of cryptic, bidirectional internal promoters at these genes. Mutational analysis confirmed that Gcn4–bound motifs within CDS can activate both sub-genic and full-length transcripts from the same or adjacent genes, demonstrating that functional Gcn4 binding is not confined to promoters. Our results show that internal promoters can be regulated by a well defined activator that also functions at conventional 5′ positioned promoters.
Click image to view.
Rps3/uS3 plays a critical role in promoting mRNA binding at the 40S entry site and in stabilizing the preinitiation complex at the start codon.
A model describing known conformational rearrangements of the PIC during scanning and start codon recognition. (i) eIF1 and the scanning enhancers (SEs) in the C-terminal tail (CTT) of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly binds. Rps3 (uS3) is located on the solvent-exposed surface of the 40S near the entry channel; the bulk of eIF3 binds on the solvent-exposed surface with a prominent domain at the mRNA exit channel; (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAiMet bound in the POUT state. eIF2 can hydrolyze GTP to GDP•Pi, but release of Pi is blocked. (iii) On AUG recognition, Met-tRNAiMet moves from the POUT to the PIN state, clashing with eIF1 and the CTT of eIF1A, provoking displacement of the eIF1A CTT from the P site, dissociation of eIF1 from the 40S subunit, and Pi release from eIF2. The N-terminal tail (NTT) of eIF1A, harboring scanning inhibitor (SI) elements, adopts a defined conformation and interacts with the codon:anticodon helix. (Above) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and impede transition to PIN state, whereas the scanning inhibitor (SI) element in the NTT of eIF1A stabilizes the PIN state. (Below) In contact with mRNA at the entry channel, uS3/Rps3 residues R116/R117 stabilize the PIN state and also promote PIC interaction with mRNA at the entry channel, augmenting the role of eIF3 in PIC-mRNA interactions at the exit channel.
Publications
- Martin-Marcos P, Zhou F, Karunasiri C, Zhang F, Dong J, Nanda J, Kulkarni SD, Sen ND, Tamame M, Zeschnigk M, Lorsch JR, Hinnebusch AG. eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast. eLife 2017;6:e31250.
- Rawal Y, Chereji RV, Valabhoju V, Qiu H, Ocampo J, Clark DJ, Hinnebusch AG. Binding in coding regions can activate internal and canonical 5' promoters in yeast. Mol Cell 2018;18:30188-30196.
- Thakur A, Hinnebusch AG. eIF1 Loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex. Proc Natl Acad Sci USA 2018;115:E4159-E4168.
- Young DJ, Makeeva DS, Zhang F, Anisimova AS, Stolboushkina EA, Ghobakhlou F, Shatsky IN, Dmitriev SE, Hinnebusch AG, Guydosh NR. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR recycle post-termination 40S subunits in vivo. Mol Cell 2018;71:761-774.
- Rawal Y, Chereji RV, Qiu H, Ananthakrishnan S, Govind CK, Clark DJ, Hinnebusch AG. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev 2018;32:695-710.
Collaborators
- David Clark, PhD, Section on Chromatin and Gene Expression, NICHD, Bethesda, MD
- Sergey E. Dmitriev, PhD, Lomonosov Moscow State University, Moscow, Russia
- Chhabi Govind, PhD, Oakland University, Rochester, MI
- Nicholas Guydosh, PhD, Section on mRNA Regulation and Translation, NIDDK, Bethesda, MD
- Nicholas Ingolia, PhD, University of California Berkeley, Berkeley, CA
- Jon Lorsch, PhD, Laboratory on the Mechanism and Regulation of Protein Synthesis, NICHD, Bethesda, MD
- Pilar Martin-Marcos, PhD, Instituto de Biologia Funcional y Genomica, Universidad de Salamanca, Salamanca, Spain
- Mercedes Tamame-González, PhD, Instituto de Biología Funcional y Genómica, Universidad de Salamanca, Salamanca, Spain
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom
- Michael Zeschnigk, PhD, Institut für Humangenetik, Universitätsklinikum, Essen, Germany
Contact
For more information, email hinnebua@mail.nih.gov or visit http://sncge.nichd.nih.gov.


